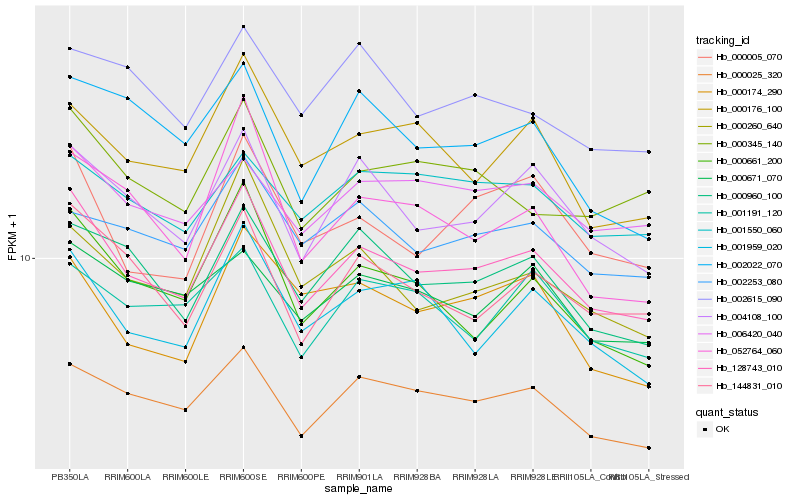
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_128743_010 |
0.0 |
- |
- |
RNA-binding protein, putative [Ricinus communis] |
| 2 |
Hb_004108_100 |
0.0603105368 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 33B isoform X2 [Jatropha curcas] |
| 3 |
Hb_000025_320 |
0.0604536634 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g28010 [Jatropha curcas] |
| 4 |
Hb_144831_010 |
0.0630546631 |
transcription factor |
TF Family: MYB |
transcription factor, putative [Ricinus communis] |
| 5 |
Hb_000960_100 |
0.068733226 |
- |
- |
PREDICTED: uncharacterized protein At1g51745 [Jatropha curcas] |
| 6 |
Hb_000661_200 |
0.0706559077 |
- |
- |
PREDICTED: uncharacterized protein LOC105648311 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001959_020 |
0.0712222541 |
- |
- |
PREDICTED: uncharacterized protein LOC105638454 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000005_070 |
0.0725312824 |
transcription factor |
TF Family: PHD |
protein binding protein, putative [Ricinus communis] |
| 9 |
Hb_000345_140 |
0.0731435405 |
- |
- |
PREDICTED: autophagy-related protein 18a-like [Jatropha curcas] |
| 10 |
Hb_000176_100 |
0.0759571894 |
- |
- |
PREDICTED: probable splicing factor 3A subunit 1 [Solanum lycopersicum] |
| 11 |
Hb_000671_070 |
0.0767688703 |
- |
- |
PREDICTED: uncharacterized protein LOC105630621 [Jatropha curcas] |
| 12 |
Hb_006420_040 |
0.0769090779 |
transcription factor |
TF Family: Orphans |
PREDICTED: paired amphipathic helix protein Sin3-like 4 isoform X2 [Jatropha curcas] |
| 13 |
Hb_001191_120 |
0.0791588098 |
- |
- |
PREDICTED: cleavage stimulating factor 64 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000260_640 |
0.0792207151 |
- |
- |
PREDICTED: uncharacterized protein At1g51745 [Jatropha curcas] |
| 15 |
Hb_002022_070 |
0.0814262334 |
- |
- |
PREDICTED: histidine--tRNA ligase [Jatropha curcas] |
| 16 |
Hb_052764_060 |
0.0814530773 |
- |
- |
XPA-binding protein, putative [Ricinus communis] |
| 17 |
Hb_002253_080 |
0.0816935611 |
- |
- |
PREDICTED: phosphatidylinositol 4-kinase beta 1-like [Jatropha curcas] |
| 18 |
Hb_001550_060 |
0.082382105 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase BSL3 isoform X4 [Jatropha curcas] |
| 19 |
Hb_002615_090 |
0.0831756297 |
- |
- |
PREDICTED: protein SMG7 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000174_290 |
0.0846902758 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 1 [Jatropha curcas] |