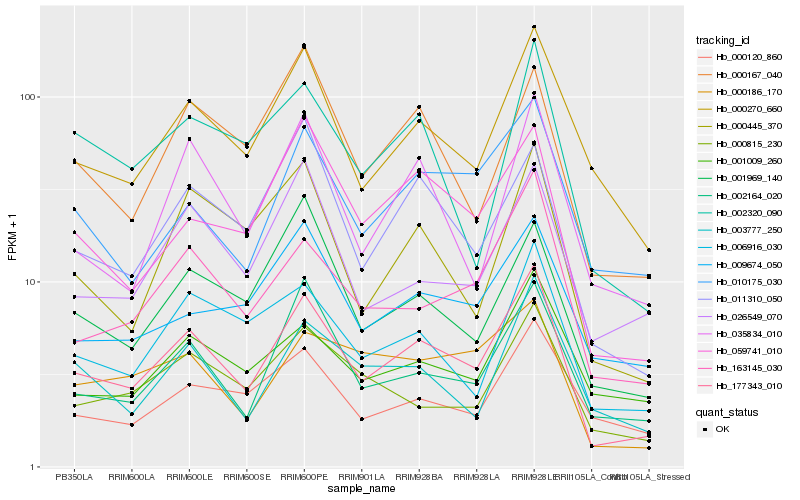
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_177343_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105650963 [Jatropha curcas] |
| 2 |
Hb_000270_660 |
0.1242792289 |
- |
- |
purine permease, putative [Ricinus communis] |
| 3 |
Hb_163145_030 |
0.1251863221 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g46580, chloroplastic [Jatropha curcas] |
| 4 |
Hb_059741_010 |
0.1260496748 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 1 [UDP-forming] [Jatropha curcas] |
| 5 |
Hb_002164_020 |
0.1280312761 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_011310_050 |
0.1357572582 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 1 [UDP-forming] [Jatropha curcas] |
| 7 |
Hb_010175_030 |
0.1370292424 |
- |
- |
PREDICTED: dentin sialophosphoprotein [Jatropha curcas] |
| 8 |
Hb_006916_030 |
0.1386103582 |
- |
- |
PREDICTED: uncharacterized protein LOC105646760 [Jatropha curcas] |
| 9 |
Hb_035834_010 |
0.1404386717 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 10 |
Hb_000445_370 |
0.1431385032 |
- |
- |
PREDICTED: probable methyltransferase PMT24 [Jatropha curcas] |
| 11 |
Hb_001969_140 |
0.14403465 |
- |
- |
PREDICTED: uncharacterized protein LOC105634929 [Jatropha curcas] |
| 12 |
Hb_000815_230 |
0.1451328436 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002320_090 |
0.1451622898 |
- |
- |
PREDICTED: endoglucanase 25-like [Jatropha curcas] |
| 14 |
Hb_003777_250 |
0.1454961403 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 2 [UDP-forming]-like [Jatropha curcas] |
| 15 |
Hb_000167_040 |
0.1471945649 |
- |
- |
PREDICTED: serine carboxypeptidase-like [Jatropha curcas] |
| 16 |
Hb_026549_070 |
0.1472931641 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 1 [Jatropha curcas] |
| 17 |
Hb_001009_260 |
0.148444898 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase 1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000186_170 |
0.1484451744 |
- |
- |
pyrophosphate-dependent phosphofructokinase, partial [Hevea brasiliensis] |
| 19 |
Hb_009674_050 |
0.1485481608 |
- |
- |
Endosomal P24A protein precursor, putative [Ricinus communis] |
| 20 |
Hb_000120_860 |
0.1487504193 |
- |
- |
nucellin, putative [Ricinus communis] |