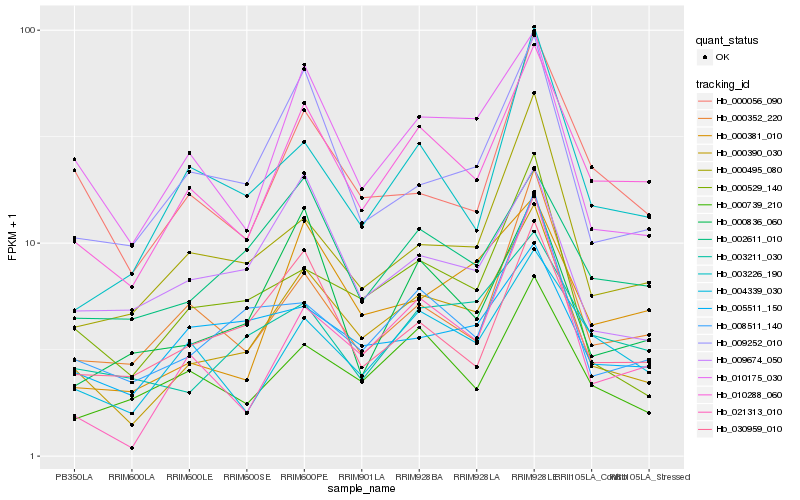
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000390_030 |
0.0 |
- |
- |
hypothetical protein ZEAMMB73_364791 [Zea mays] |
| 2 |
Hb_000529_140 |
0.1171335428 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_010288_060 |
0.1220544412 |
- |
- |
PREDICTED: dnaJ protein ERDJ3B [Jatropha curcas] |
| 4 |
Hb_010175_030 |
0.1266702997 |
- |
- |
PREDICTED: dentin sialophosphoprotein [Jatropha curcas] |
| 5 |
Hb_008511_140 |
0.1360710234 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105632314 [Jatropha curcas] |
| 6 |
Hb_009252_010 |
0.1429871939 |
- |
- |
PREDICTED: kinesin light chain [Jatropha curcas] |
| 7 |
Hb_000495_080 |
0.1486998704 |
- |
- |
Pentatricopeptide repeat (PPR-like) superfamily protein [Theobroma cacao] |
| 8 |
Hb_021313_010 |
0.1544415209 |
- |
- |
hypothetical protein POPTR_0010s24380g [Populus trichocarpa] |
| 9 |
Hb_000739_210 |
0.1560484934 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g01970 [Jatropha curcas] |
| 10 |
Hb_030959_010 |
0.1560772548 |
- |
- |
cullin, putative [Ricinus communis] |
| 11 |
Hb_000381_010 |
0.1561699429 |
- |
- |
PREDICTED: arogenate dehydratase/prephenate dehydratase 1, chloroplastic-like [Jatropha curcas] |
| 12 |
Hb_003211_030 |
0.1589484041 |
- |
- |
- |
| 13 |
Hb_009674_050 |
0.1603604039 |
- |
- |
Endosomal P24A protein precursor, putative [Ricinus communis] |
| 14 |
Hb_000352_220 |
0.1619330911 |
- |
- |
PREDICTED: probable tyrosine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 15 |
Hb_005511_150 |
0.163342007 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g71810, chloroplastic isoform X5 [Populus euphratica] |
| 16 |
Hb_000836_060 |
0.1642741779 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-2 [Jatropha curcas] |
| 17 |
Hb_000056_090 |
0.1650719638 |
- |
- |
UDP-galactose transporter 3 isoform 1 [Theobroma cacao] |
| 18 |
Hb_002611_010 |
0.1655173494 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 19 |
Hb_004339_030 |
0.1659913667 |
- |
- |
PREDICTED: 7-methylguanosine phosphate-specific 5'-nucleotidase A isoform X2 [Jatropha curcas] |
| 20 |
Hb_003226_190 |
0.1663448881 |
- |
- |
PREDICTED: blue-light photoreceptor PHR2 [Jatropha curcas] |