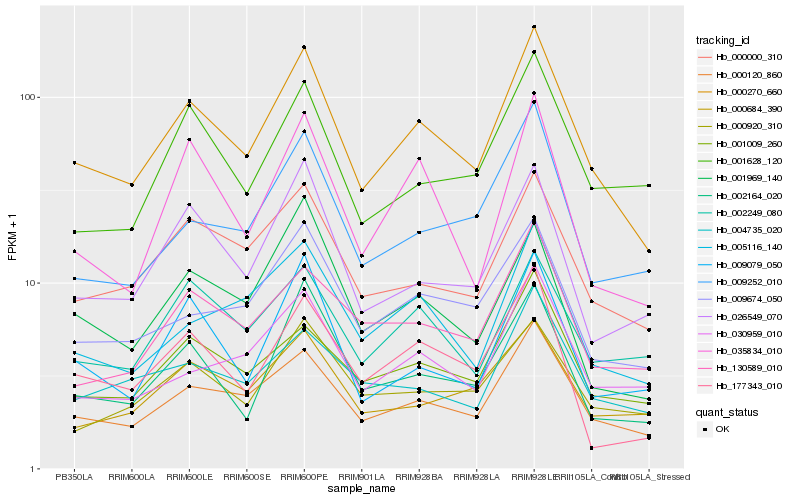
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000270_660 |
0.0 |
- |
- |
purine permease, putative [Ricinus communis] |
| 2 |
Hb_000120_860 |
0.0709005891 |
- |
- |
nucellin, putative [Ricinus communis] |
| 3 |
Hb_000000_310 |
0.1008210233 |
- |
- |
metalloendopeptidase, putative [Ricinus communis] |
| 4 |
Hb_001009_260 |
0.1092895497 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase 1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_009674_050 |
0.1103228684 |
- |
- |
Endosomal P24A protein precursor, putative [Ricinus communis] |
| 6 |
Hb_002164_020 |
0.1119635415 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_005116_140 |
0.1133348344 |
- |
- |
PREDICTED: RAN GTPase-activating protein 1 [Jatropha curcas] |
| 8 |
Hb_026549_070 |
0.1157844159 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 1 [Jatropha curcas] |
| 9 |
Hb_002249_080 |
0.1192379461 |
- |
- |
lysosomal alpha-mannosidase, putative [Ricinus communis] |
| 10 |
Hb_030959_010 |
0.1197970935 |
- |
- |
cullin, putative [Ricinus communis] |
| 11 |
Hb_009079_050 |
0.1207778166 |
- |
- |
PREDICTED: V-type proton ATPase subunit a3-like [Jatropha curcas] |
| 12 |
Hb_009252_010 |
0.1207882584 |
- |
- |
PREDICTED: kinesin light chain [Jatropha curcas] |
| 13 |
Hb_035834_010 |
0.1212581154 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 14 |
Hb_001969_140 |
0.1225368017 |
- |
- |
PREDICTED: uncharacterized protein LOC105634929 [Jatropha curcas] |
| 15 |
Hb_000920_310 |
0.1225791711 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 16 |
Hb_177343_010 |
0.1242792289 |
- |
- |
PREDICTED: uncharacterized protein LOC105650963 [Jatropha curcas] |
| 17 |
Hb_130589_010 |
0.124427187 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 18 |
Hb_004735_020 |
0.1252956833 |
- |
- |
PREDICTED: uncharacterized protein LOC105639315 [Jatropha curcas] |
| 19 |
Hb_000684_390 |
0.1255080152 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 4 [Jatropha curcas] |
| 20 |
Hb_001628_120 |
0.1264189054 |
- |
- |
putative ascorbate peroxidase, partial [Taraxacum brevicorniculatum] |