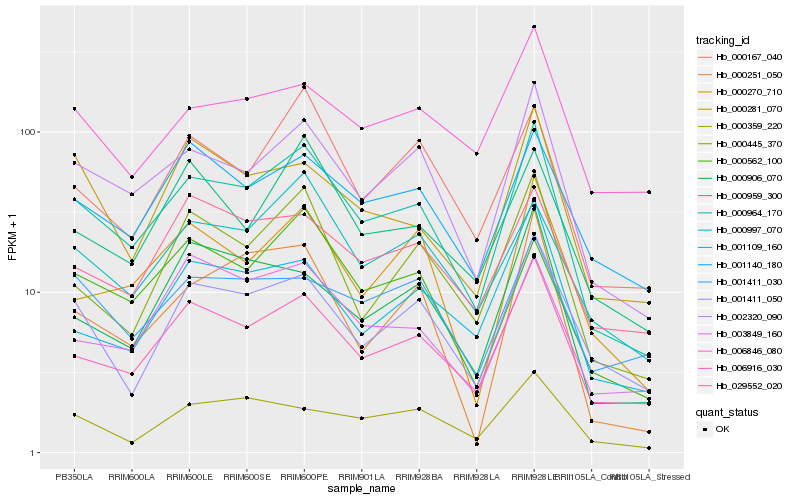
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000964_170 |
0.0 |
- |
- |
Cellulose synthase A catalytic subunit 6 [UDP-forming], putative [Ricinus communis] |
| 2 |
Hb_002320_090 |
0.0743867542 |
- |
- |
PREDICTED: endoglucanase 25-like [Jatropha curcas] |
| 3 |
Hb_006916_030 |
0.0926268715 |
- |
- |
PREDICTED: uncharacterized protein LOC105646760 [Jatropha curcas] |
| 4 |
Hb_000445_370 |
0.1228349202 |
- |
- |
PREDICTED: probable methyltransferase PMT24 [Jatropha curcas] |
| 5 |
Hb_001411_050 |
0.1228893853 |
- |
- |
PREDICTED: uncharacterized protein LOC105640956 [Jatropha curcas] |
| 6 |
Hb_001140_180 |
0.1255194363 |
- |
- |
PREDICTED: transmembrane ascorbate ferrireductase 1 [Jatropha curcas] |
| 7 |
Hb_000270_710 |
0.1300326516 |
- |
- |
PREDICTED: WAT1-related protein At5g40240-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_000997_070 |
0.1302782714 |
- |
- |
PREDICTED: ureidoglycolate hydrolase isoform X1 [Jatropha curcas] |
| 9 |
Hb_000562_100 |
0.1315146224 |
- |
- |
PREDICTED: protein IQ-DOMAIN 32 [Jatropha curcas] |
| 10 |
Hb_000167_040 |
0.1337327669 |
- |
- |
PREDICTED: serine carboxypeptidase-like [Jatropha curcas] |
| 11 |
Hb_000959_300 |
0.1366293971 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 2 [UDP-forming] isoform X2 [Jatropha curcas] |
| 12 |
Hb_000906_070 |
0.1371123637 |
- |
- |
PREDICTED: protein SCAR2-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_000251_050 |
0.1378080382 |
- |
- |
hypothetical protein JCGZ_21678 [Jatropha curcas] |
| 14 |
Hb_000359_220 |
0.1379861266 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g42920, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000281_070 |
0.1389278276 |
- |
- |
rubisco subunit binding-protein beta subunit, rubb, putative [Ricinus communis] |
| 16 |
Hb_001109_160 |
0.1395138523 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 17 |
Hb_029552_020 |
0.1398795318 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_003849_160 |
0.1398828931 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase XERICO [Jatropha curcas] |
| 19 |
Hb_006846_080 |
0.1402596625 |
- |
- |
calnexin, putative [Ricinus communis] |
| 20 |
Hb_001411_030 |
0.1447098846 |
- |
- |
hypothetical protein POPTR_0003s09620g [Populus trichocarpa] |