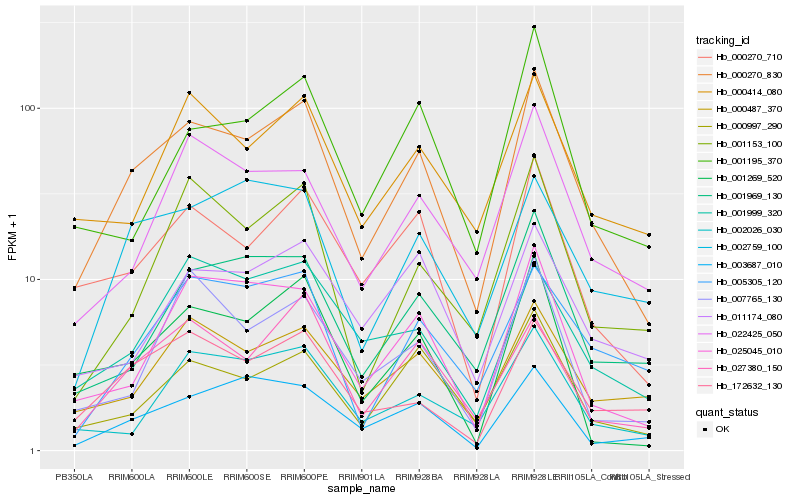
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000270_830 |
0.0 |
- |
- |
aldehyde dehydrogenase, putative [Ricinus communis] |
| 2 |
Hb_000270_710 |
0.1252736884 |
- |
- |
PREDICTED: WAT1-related protein At5g40240-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_025045_010 |
0.1374250183 |
- |
- |
catalytic, putative [Ricinus communis] |
| 4 |
Hb_000997_290 |
0.1383707226 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme [Jatropha curcas] |
| 5 |
Hb_027380_150 |
0.143257915 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001969_130 |
0.1436480952 |
- |
- |
PREDICTED: molybdate transporter 2 [Jatropha curcas] |
| 7 |
Hb_000487_370 |
0.1459088126 |
- |
- |
- |
| 8 |
Hb_022425_050 |
0.1476664466 |
- |
- |
PREDICTED: probable lactoylglutathione lyase, chloroplast isoform X1 [Jatropha curcas] |
| 9 |
Hb_000414_080 |
0.1505629363 |
- |
- |
PREDICTED: triosephosphate isomerase, chloroplastic [Jatropha curcas] |
| 10 |
Hb_001269_520 |
0.1515697037 |
- |
- |
PREDICTED: putative receptor-like protein kinase At1g80870 [Jatropha curcas] |
| 11 |
Hb_172632_130 |
0.1525693267 |
- |
- |
hypothetical protein JCGZ_22584 [Jatropha curcas] |
| 12 |
Hb_003687_010 |
0.1529999398 |
- |
- |
Cysteine-rich RLK 29, putative [Theobroma cacao] |
| 13 |
Hb_002026_030 |
0.1555172488 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 14 |
Hb_001153_100 |
0.156267308 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein HD1 [Jatropha curcas] |
| 15 |
Hb_002759_100 |
0.1580057144 |
- |
- |
hypothetical protein JCGZ_21777 [Jatropha curcas] |
| 16 |
Hb_001195_370 |
0.1596412707 |
- |
- |
- |
| 17 |
Hb_001999_320 |
0.1612196778 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor DIVARICATA [Jatropha curcas] |
| 18 |
Hb_011174_080 |
0.1628485815 |
- |
- |
PREDICTED: sulfite oxidase [Jatropha curcas] |
| 19 |
Hb_005305_120 |
0.1632697221 |
- |
- |
PREDICTED: calcium-dependent protein kinase 8-like [Jatropha curcas] |
| 20 |
Hb_007765_130 |
0.1633996784 |
- |
- |
PREDICTED: UPF0503 protein At3g09070, chloroplastic [Jatropha curcas] |