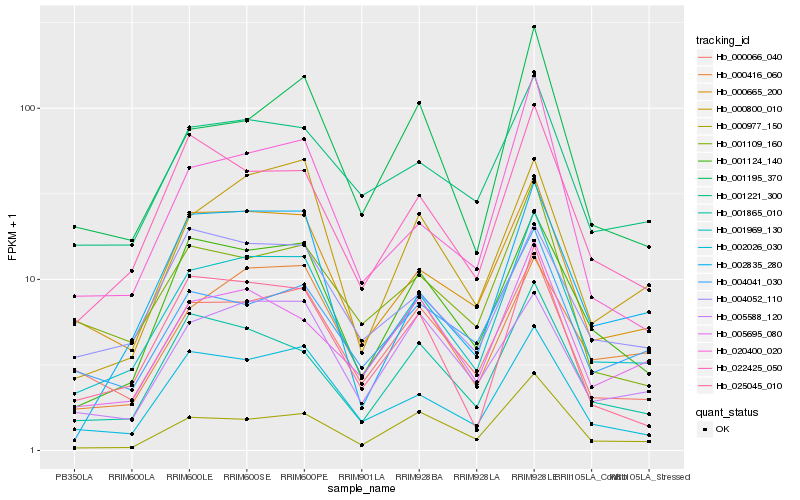
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001969_130 |
0.0 |
- |
- |
PREDICTED: molybdate transporter 2 [Jatropha curcas] |
| 2 |
Hb_000665_200 |
0.0876625276 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 3 |
Hb_001124_140 |
0.0929751699 |
- |
- |
PREDICTED: inactive beta-amylase 9 [Jatropha curcas] |
| 4 |
Hb_002026_030 |
0.0943275038 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 5 |
Hb_022425_050 |
0.0997439539 |
- |
- |
PREDICTED: probable lactoylglutathione lyase, chloroplast isoform X1 [Jatropha curcas] |
| 6 |
Hb_000066_040 |
0.1032304382 |
- |
- |
PREDICTED: CDPK-related kinase 7-like [Populus euphratica] |
| 7 |
Hb_001221_300 |
0.1064957718 |
- |
- |
PREDICTED: acetolactate synthase 3, chloroplastic [Jatropha curcas] |
| 8 |
Hb_001865_010 |
0.107225844 |
transcription factor |
TF Family: B3 |
hypothetical protein RCOM_0465210 [Ricinus communis] |
| 9 |
Hb_005695_080 |
0.1120592256 |
- |
- |
PREDICTED: acetyl-CoA carboxylase 1-like [Jatropha curcas] |
| 10 |
Hb_025045_010 |
0.1202847813 |
- |
- |
catalytic, putative [Ricinus communis] |
| 11 |
Hb_000416_060 |
0.1204608139 |
- |
- |
PREDICTED: uncharacterized protein LOC105649718 [Jatropha curcas] |
| 12 |
Hb_004041_030 |
0.1228019186 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein C-like isoform X1 [Populus euphratica] |
| 13 |
Hb_001195_370 |
0.1232944037 |
- |
- |
- |
| 14 |
Hb_000800_010 |
0.1240235226 |
- |
- |
PREDICTED: glycosyltransferase family protein 64 protein C5 [Jatropha curcas] |
| 15 |
Hb_004052_110 |
0.1250289968 |
- |
- |
PREDICTED: host cell factor 2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_020400_020 |
0.1255804383 |
- |
- |
starch synthase isoform II [Manihot esculenta] |
| 17 |
Hb_000977_150 |
0.1276188854 |
- |
- |
latex cyanogenic beta glucosidase [Hevea brasiliensis] |
| 18 |
Hb_005588_120 |
0.1278190069 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001109_160 |
0.129267167 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 20 |
Hb_002835_280 |
0.1294603382 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase S.1 [Jatropha curcas] |