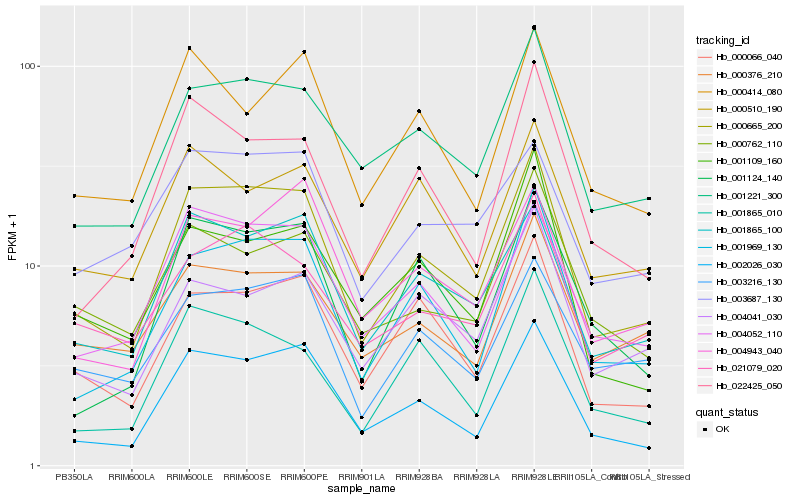
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000665_200 |
0.0 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 2 |
Hb_002026_030 |
0.0827565848 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 3 |
Hb_001865_100 |
0.0860352705 |
- |
- |
PREDICTED: ACT domain-containing protein ACR9 [Jatropha curcas] |
| 4 |
Hb_001969_130 |
0.0876625276 |
- |
- |
PREDICTED: molybdate transporter 2 [Jatropha curcas] |
| 5 |
Hb_004052_110 |
0.0936125074 |
- |
- |
PREDICTED: host cell factor 2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001221_300 |
0.0937900105 |
- |
- |
PREDICTED: acetolactate synthase 3, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000066_040 |
0.095188936 |
- |
- |
PREDICTED: CDPK-related kinase 7-like [Populus euphratica] |
| 8 |
Hb_003216_130 |
0.1079132463 |
- |
- |
PREDICTED: uncharacterized protein LOC105641500 [Jatropha curcas] |
| 9 |
Hb_021079_020 |
0.1095798536 |
- |
- |
PREDICTED: uncharacterized protein LOC105648580 [Jatropha curcas] |
| 10 |
Hb_004943_040 |
0.1130204983 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g30440 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001124_140 |
0.1138511066 |
- |
- |
PREDICTED: inactive beta-amylase 9 [Jatropha curcas] |
| 12 |
Hb_022425_050 |
0.114347996 |
- |
- |
PREDICTED: probable lactoylglutathione lyase, chloroplast isoform X1 [Jatropha curcas] |
| 13 |
Hb_000376_210 |
0.1180608066 |
- |
- |
PREDICTED: O-acyltransferase WSD1-like [Jatropha curcas] |
| 14 |
Hb_001865_010 |
0.1196287913 |
transcription factor |
TF Family: B3 |
hypothetical protein RCOM_0465210 [Ricinus communis] |
| 15 |
Hb_000414_080 |
0.1212442633 |
- |
- |
PREDICTED: triosephosphate isomerase, chloroplastic [Jatropha curcas] |
| 16 |
Hb_004041_030 |
0.1213746115 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein C-like isoform X1 [Populus euphratica] |
| 17 |
Hb_000762_110 |
0.121631402 |
- |
- |
PREDICTED: protease Do-like 1, chloroplastic [Jatropha curcas] |
| 18 |
Hb_001109_160 |
0.1226811075 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 19 |
Hb_003687_130 |
0.1232238731 |
- |
- |
PREDICTED: probable methyltransferase PMT3 [Jatropha curcas] |
| 20 |
Hb_000510_190 |
0.1238751968 |
- |
- |
glutathione reductase [Hevea brasiliensis] |