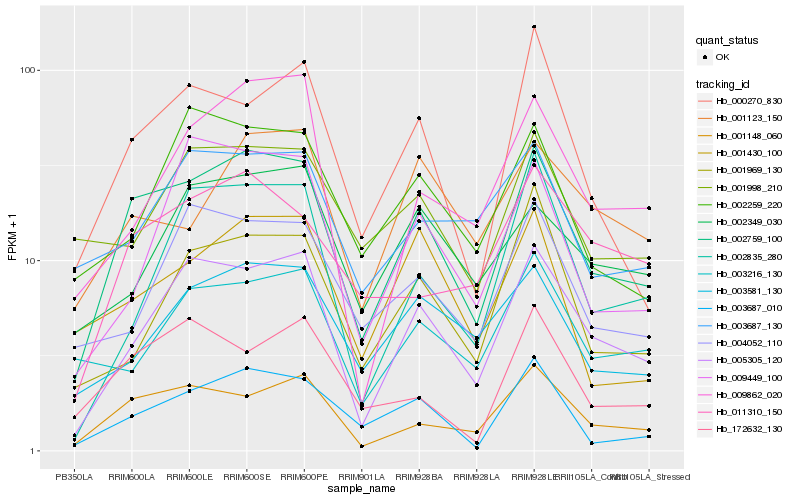
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002759_100 |
0.0 |
- |
- |
hypothetical protein JCGZ_21777 [Jatropha curcas] |
| 2 |
Hb_005305_120 |
0.1237884687 |
- |
- |
PREDICTED: calcium-dependent protein kinase 8-like [Jatropha curcas] |
| 3 |
Hb_001148_060 |
0.1273207383 |
- |
- |
hypothetical protein JCGZ_00278 [Jatropha curcas] |
| 4 |
Hb_003687_010 |
0.1446295322 |
- |
- |
Cysteine-rich RLK 29, putative [Theobroma cacao] |
| 5 |
Hb_000270_830 |
0.1580057144 |
- |
- |
aldehyde dehydrogenase, putative [Ricinus communis] |
| 6 |
Hb_003581_130 |
0.161595931 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 7 |
Hb_011310_150 |
0.1696521355 |
- |
- |
PREDICTED: SPX domain-containing protein 1-like [Jatropha curcas] |
| 8 |
Hb_004052_110 |
0.1699389102 |
- |
- |
PREDICTED: host cell factor 2 isoform X1 [Jatropha curcas] |
| 9 |
Hb_172632_130 |
0.1705847685 |
- |
- |
hypothetical protein JCGZ_22584 [Jatropha curcas] |
| 10 |
Hb_001430_100 |
0.1709495286 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 11 |
Hb_002259_220 |
0.171581119 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 66 [Jatropha curcas] |
| 12 |
Hb_009862_020 |
0.1735126502 |
- |
- |
PREDICTED: uncharacterized protein LOC105650670 [Jatropha curcas] |
| 13 |
Hb_001969_130 |
0.1742386466 |
- |
- |
PREDICTED: molybdate transporter 2 [Jatropha curcas] |
| 14 |
Hb_001998_210 |
0.1744034026 |
- |
- |
unnamed protein product [Coffea canephora] |
| 15 |
Hb_009449_100 |
0.1762297657 |
- |
- |
PREDICTED: equilibrative nucleotide transporter 1 [Jatropha curcas] |
| 16 |
Hb_003687_130 |
0.1762718446 |
- |
- |
PREDICTED: probable methyltransferase PMT3 [Jatropha curcas] |
| 17 |
Hb_001123_150 |
0.1764555434 |
transcription factor |
TF Family: NAC |
NAC transcription factor [Hevea brasiliensis] |
| 18 |
Hb_003216_130 |
0.176732631 |
- |
- |
PREDICTED: uncharacterized protein LOC105641500 [Jatropha curcas] |
| 19 |
Hb_002835_280 |
0.1772857708 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase S.1 [Jatropha curcas] |
| 20 |
Hb_002349_030 |
0.1789579499 |
- |
- |
conserved hypothetical protein [Ricinus communis] |