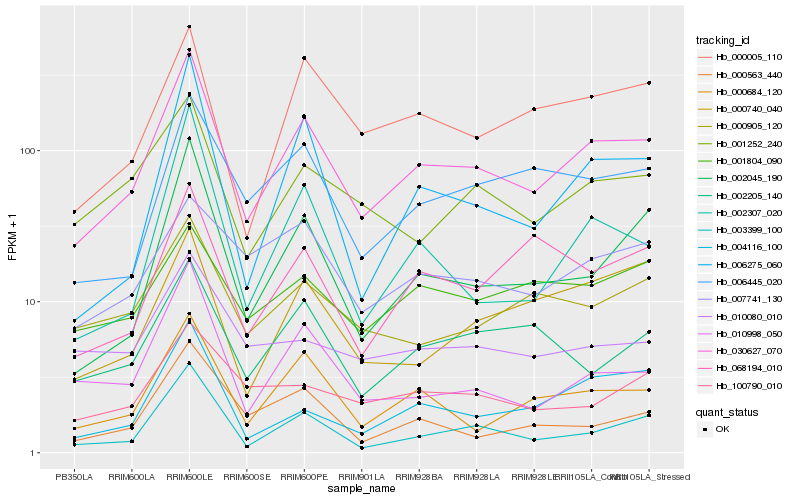
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_030627_070 |
0.0 |
- |
- |
histone H4 [Zea mays] |
| 2 |
Hb_003399_100 |
0.1202315565 |
- |
- |
PREDICTED: putative cyclin-A3-1 [Jatropha curcas] |
| 3 |
Hb_000684_120 |
0.1233938327 |
- |
- |
PREDICTED: uncharacterized protein LOC105642190 isoform X2 [Jatropha curcas] |
| 4 |
Hb_002045_190 |
0.1354872877 |
- |
- |
PREDICTED: histone H2A variant 1 [Jatropha curcas] |
| 5 |
Hb_000005_110 |
0.1376691585 |
- |
- |
histone H2B1 [Hevea brasiliensis] |
| 6 |
Hb_006275_060 |
0.1387858809 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 7 |
Hb_006445_020 |
0.1437156358 |
- |
- |
hypothetical protein L484_026741 [Morus notabilis] |
| 8 |
Hb_010998_050 |
0.147235063 |
- |
- |
PREDICTED: kinesin-like protein KIF22 isoform X2 [Jatropha curcas] |
| 9 |
Hb_001252_240 |
0.1476416956 |
- |
- |
PREDICTED: protein SPIRAL1-like 3 [Jatropha curcas] |
| 10 |
Hb_000563_440 |
0.1511567922 |
- |
- |
PREDICTED: condensin-2 complex subunit H2 isoform X2 [Jatropha curcas] |
| 11 |
Hb_004116_100 |
0.1558841945 |
- |
- |
Isoamyl acetate-hydrolyzing esterase, putative [Ricinus communis] |
| 12 |
Hb_002205_140 |
0.1583640391 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor RF2b [Jatropha curcas] |
| 13 |
Hb_000905_120 |
0.1596948631 |
- |
- |
PREDICTED: uncharacterized protein LOC105628677 [Jatropha curcas] |
| 14 |
Hb_002307_020 |
0.1611469497 |
- |
- |
PREDICTED: histone H1-like [Jatropha curcas] |
| 15 |
Hb_068194_010 |
0.1638508137 |
- |
- |
hypothetical protein POPTR_0010s23740g [Populus trichocarpa] |
| 16 |
Hb_000740_040 |
0.1675793482 |
- |
- |
PREDICTED: uncharacterized protein LOC105126082 [Populus euphratica] |
| 17 |
Hb_100790_010 |
0.168923939 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_007741_130 |
0.1690598919 |
- |
- |
hypothetical protein B456_007G089000 [Gossypium raimondii] |
| 19 |
Hb_010080_010 |
0.171443097 |
- |
- |
PREDICTED: uncharacterized protein LOC105635247 [Jatropha curcas] |
| 20 |
Hb_001804_090 |
0.1747656588 |
- |
- |
PREDICTED: uracil phosphoribosyltransferase isoform X1 [Jatropha curcas] |