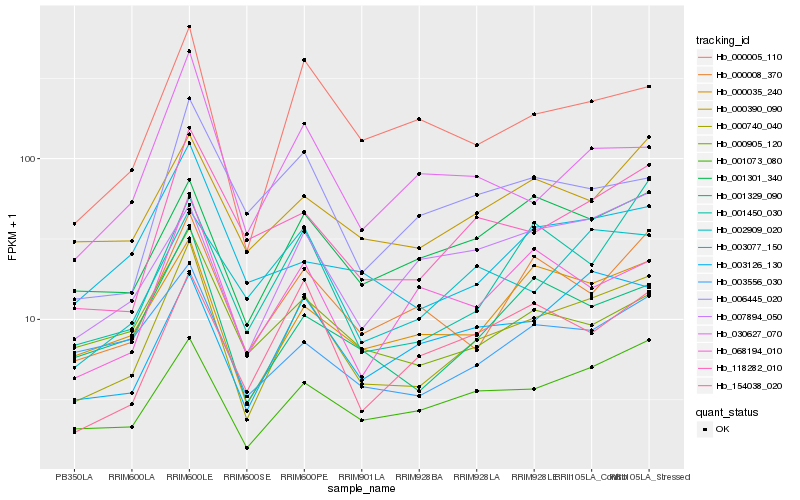
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000740_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105126082 [Populus euphratica] |
| 2 |
Hb_001073_080 |
0.1189164669 |
- |
- |
PREDICTED: protein root UVB sensitive 2, chloroplastic [Jatropha curcas] |
| 3 |
Hb_118282_010 |
0.1357183484 |
- |
- |
PREDICTED: uncharacterized protein LOC105108358 [Populus euphratica] |
| 4 |
Hb_001329_090 |
0.1382477757 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000005_110 |
0.1384321172 |
- |
- |
histone H2B1 [Hevea brasiliensis] |
| 6 |
Hb_000035_240 |
0.1403111422 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003126_130 |
0.1448098539 |
- |
- |
inositol-1-monophosphatase family protein [Populus trichocarpa] |
| 8 |
Hb_000008_370 |
0.1465483717 |
- |
- |
PREDICTED: uncharacterized protein LOC105628095 [Jatropha curcas] |
| 9 |
Hb_001301_340 |
0.1491580849 |
- |
- |
endoribonuclease L-PSP family protein [Populus trichocarpa] |
| 10 |
Hb_002909_020 |
0.1521715575 |
- |
- |
PREDICTED: uncharacterized protein LOC105632476 [Jatropha curcas] |
| 11 |
Hb_154038_020 |
0.1530085706 |
- |
- |
hypothetical protein JCGZ_05648 [Jatropha curcas] |
| 12 |
Hb_000390_090 |
0.1535767465 |
- |
- |
PREDICTED: uncharacterized protein LOC105632476 [Jatropha curcas] |
| 13 |
Hb_068194_010 |
0.1565914898 |
- |
- |
hypothetical protein POPTR_0010s23740g [Populus trichocarpa] |
| 14 |
Hb_007894_050 |
0.1571282767 |
- |
- |
PREDICTED: reactive Intermediate Deaminase A, chloroplastic [Jatropha curcas] |
| 15 |
Hb_003556_030 |
0.1612124463 |
- |
- |
PREDICTED: uncharacterized oxidoreductase C663.09c isoform X1 [Jatropha curcas] |
| 16 |
Hb_001450_030 |
0.1614879613 |
- |
- |
PREDICTED: uncharacterized protein LOC105650827 [Jatropha curcas] |
| 17 |
Hb_000905_120 |
0.1643010287 |
- |
- |
PREDICTED: uncharacterized protein LOC105628677 [Jatropha curcas] |
| 18 |
Hb_003077_150 |
0.1651027259 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
brg-1 associated factor, putative [Ricinus communis] |
| 19 |
Hb_030627_070 |
0.1675793482 |
- |
- |
histone H4 [Zea mays] |
| 20 |
Hb_006445_020 |
0.1679366452 |
- |
- |
hypothetical protein L484_026741 [Morus notabilis] |