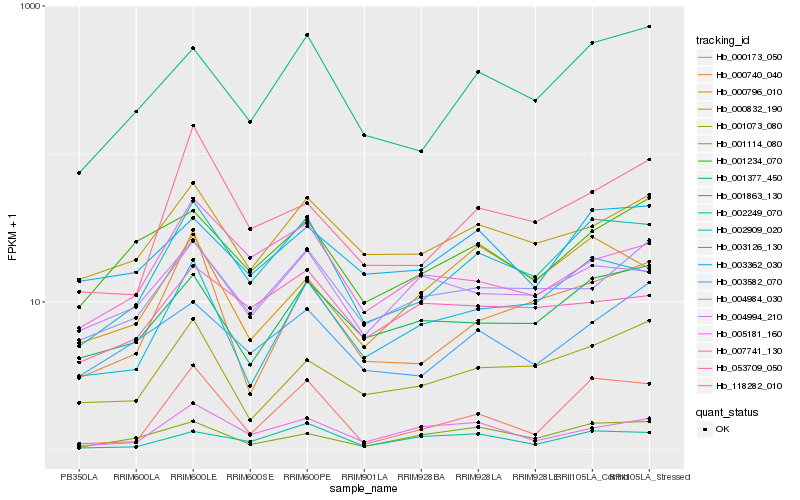
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002909_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632476 [Jatropha curcas] |
| 2 |
Hb_001863_130 |
0.1036679259 |
- |
- |
hypothetical protein ZEAMMB73_433257, partial [Zea mays] |
| 3 |
Hb_003126_130 |
0.1248023539 |
- |
- |
inositol-1-monophosphatase family protein [Populus trichocarpa] |
| 4 |
Hb_000832_190 |
0.1299072404 |
- |
- |
Rab3 [Hevea brasiliensis] |
| 5 |
Hb_001377_450 |
0.1310755146 |
- |
- |
PREDICTED: pyridoxine/pyridoxamine 5'-phosphate oxidase 2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_004984_030 |
0.1367663496 |
- |
- |
protein with unknown function [Ricinus communis] |
| 7 |
Hb_007741_130 |
0.1382055162 |
- |
- |
hypothetical protein B456_007G089000 [Gossypium raimondii] |
| 8 |
Hb_004994_210 |
0.1420776432 |
- |
- |
phosphomannomutase family protein [Populus trichocarpa] |
| 9 |
Hb_001234_070 |
0.1449807451 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein Sgpp [Populus euphratica] |
| 10 |
Hb_003582_070 |
0.1461594898 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_003362_030 |
0.1467050215 |
- |
- |
hypothetical protein POPTR_0017s08440g [Populus trichocarpa] |
| 12 |
Hb_005181_160 |
0.1470197743 |
- |
- |
PREDICTED: uncharacterized protein LOC105643546 [Jatropha curcas] |
| 13 |
Hb_118282_010 |
0.1471386522 |
- |
- |
PREDICTED: uncharacterized protein LOC105108358 [Populus euphratica] |
| 14 |
Hb_002249_070 |
0.1520146901 |
- |
- |
gcn4-complementing protein, putative [Ricinus communis] |
| 15 |
Hb_000740_040 |
0.1521715575 |
- |
- |
PREDICTED: uncharacterized protein LOC105126082 [Populus euphratica] |
| 16 |
Hb_001073_080 |
0.1529018904 |
- |
- |
PREDICTED: protein root UVB sensitive 2, chloroplastic [Jatropha curcas] |
| 17 |
Hb_053709_050 |
0.1549072843 |
- |
- |
PREDICTED: nudix hydrolase 23, chloroplastic isoform X2 [Jatropha curcas] |
| 18 |
Hb_000173_050 |
0.1553731299 |
- |
- |
basic 7S globulin 2 precursor small subunit, putative [Ricinus communis] |
| 19 |
Hb_000796_010 |
0.1575189967 |
- |
- |
PREDICTED: nitrogen regulatory protein P-II homolog [Jatropha curcas] |
| 20 |
Hb_001114_080 |
0.158034752 |
- |
- |
hypothetical protein PRUPE_ppa019021mg, partial [Prunus persica] |