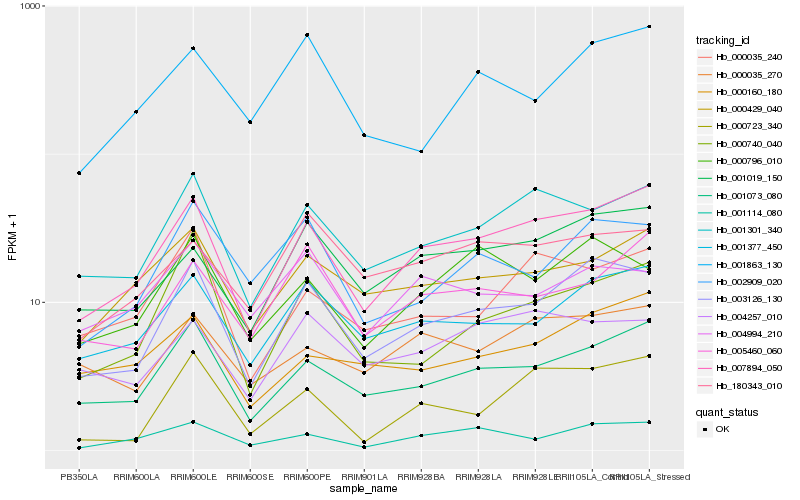
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003126_130 |
0.0 |
- |
- |
inositol-1-monophosphatase family protein [Populus trichocarpa] |
| 2 |
Hb_007894_050 |
0.1007801007 |
- |
- |
PREDICTED: reactive Intermediate Deaminase A, chloroplastic [Jatropha curcas] |
| 3 |
Hb_001377_450 |
0.1118480312 |
- |
- |
PREDICTED: pyridoxine/pyridoxamine 5'-phosphate oxidase 2 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001073_080 |
0.1125537782 |
- |
- |
PREDICTED: protein root UVB sensitive 2, chloroplastic [Jatropha curcas] |
| 5 |
Hb_002909_020 |
0.1248023539 |
- |
- |
PREDICTED: uncharacterized protein LOC105632476 [Jatropha curcas] |
| 6 |
Hb_001019_150 |
0.1315792285 |
- |
- |
Acyl-protein thioesterase, putative [Ricinus communis] |
| 7 |
Hb_000796_010 |
0.1327992004 |
- |
- |
PREDICTED: nitrogen regulatory protein P-II homolog [Jatropha curcas] |
| 8 |
Hb_001301_340 |
0.1328959714 |
- |
- |
endoribonuclease L-PSP family protein [Populus trichocarpa] |
| 9 |
Hb_000740_040 |
0.1448098539 |
- |
- |
PREDICTED: uncharacterized protein LOC105126082 [Populus euphratica] |
| 10 |
Hb_001114_080 |
0.1480995675 |
- |
- |
hypothetical protein PRUPE_ppa019021mg, partial [Prunus persica] |
| 11 |
Hb_001863_130 |
0.1527130744 |
- |
- |
hypothetical protein ZEAMMB73_433257, partial [Zea mays] |
| 12 |
Hb_000429_040 |
0.1556045947 |
- |
- |
PREDICTED: chaperone protein dnaJ 6 [Vitis vinifera] |
| 13 |
Hb_180343_010 |
0.1583053979 |
- |
- |
Biotin carboxyl carrier protein of acetyl-CoA carboxylase, putative [Ricinus communis] |
| 14 |
Hb_004257_010 |
0.1584856583 |
- |
- |
CMP-sialic acid transporter, putative [Ricinus communis] |
| 15 |
Hb_000035_270 |
0.160653641 |
- |
- |
PREDICTED: uncharacterized protein LOC105637542 [Jatropha curcas] |
| 16 |
Hb_005460_060 |
0.1622297555 |
- |
- |
PREDICTED: proteasome assembly chaperone 2 [Jatropha curcas] |
| 17 |
Hb_004994_210 |
0.1627928097 |
- |
- |
phosphomannomutase family protein [Populus trichocarpa] |
| 18 |
Hb_000160_180 |
0.1634290448 |
- |
- |
methionyl-tRNA formyltransferase, putative [Ricinus communis] |
| 19 |
Hb_000035_240 |
0.1634807661 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000723_340 |
0.1651400982 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG3 [Jatropha curcas] |