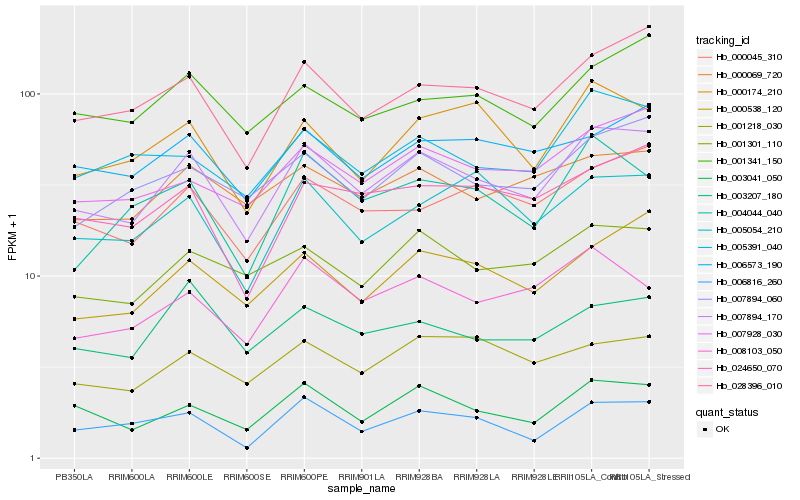
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007894_170 |
0.0 |
- |
- |
PREDICTED: UPF0136 membrane protein At2g26240-like [Jatropha curcas] |
| 2 |
Hb_028396_010 |
0.0765726598 |
- |
- |
PREDICTED: probable ATP synthase 24 kDa subunit, mitochondrial [Vitis vinifera] |
| 3 |
Hb_003041_050 |
0.0802249888 |
- |
- |
- |
| 4 |
Hb_000538_120 |
0.084468244 |
transcription factor |
TF Family: SET |
PREDICTED: protein SET DOMAIN GROUP 40 [Jatropha curcas] |
| 5 |
Hb_000045_310 |
0.0870466864 |
- |
- |
PREDICTED: probable signal peptidase complex subunit 2 [Jatropha curcas] |
| 6 |
Hb_001301_110 |
0.0881609425 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 22 homolog 1 [Jatropha curcas] |
| 7 |
Hb_007894_060 |
0.0882403641 |
- |
- |
PREDICTED: ubiquitin thioesterase OTU1 [Jatropha curcas] |
| 8 |
Hb_007928_030 |
0.088900786 |
- |
- |
PREDICTED: dual specificity phosphatase Cdc25 [Jatropha curcas] |
| 9 |
Hb_005391_040 |
0.0926487725 |
- |
- |
PREDICTED: 1,4-dihydroxy-2-naphthoyl-CoA thioesterase 1 [Jatropha curcas] |
| 10 |
Hb_024650_070 |
0.0936108466 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 7 homolog A-like [Jatropha curcas] |
| 11 |
Hb_000069_720 |
0.0940589725 |
- |
- |
PREDICTED: exosome complex component RRP41-like [Jatropha curcas] |
| 12 |
Hb_006573_190 |
0.0960583781 |
- |
- |
PREDICTED: probable NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 5, mitochondrial [Jatropha curcas] |
| 13 |
Hb_001341_150 |
0.0968075025 |
- |
- |
PREDICTED: proteasome subunit beta type-4-like [Jatropha curcas] |
| 14 |
Hb_000174_210 |
0.0972002122 |
- |
- |
latex cyanogenic beta glucosidase [Hevea brasiliensis] |
| 15 |
Hb_008103_050 |
0.0982829908 |
- |
- |
hypothetical protein JCGZ_07422 [Jatropha curcas] |
| 16 |
Hb_003207_180 |
0.0994510708 |
- |
- |
PREDICTED: uncharacterized protein LOC105645887 isoform X2 [Jatropha curcas] |
| 17 |
Hb_004044_040 |
0.0997396444 |
- |
- |
PREDICTED: NADH-cytochrome b5 reductase-like protein [Jatropha curcas] |
| 18 |
Hb_005054_210 |
0.0999285165 |
- |
- |
hypothetical protein PRUPE_ppa026456mg [Prunus persica] |
| 19 |
Hb_006816_260 |
0.1004156464 |
- |
- |
hypothetical protein JCGZ_23244 [Jatropha curcas] |
| 20 |
Hb_001218_030 |
0.1004630868 |
- |
- |
PREDICTED: uncharacterized protein LOC105630612 [Jatropha curcas] |