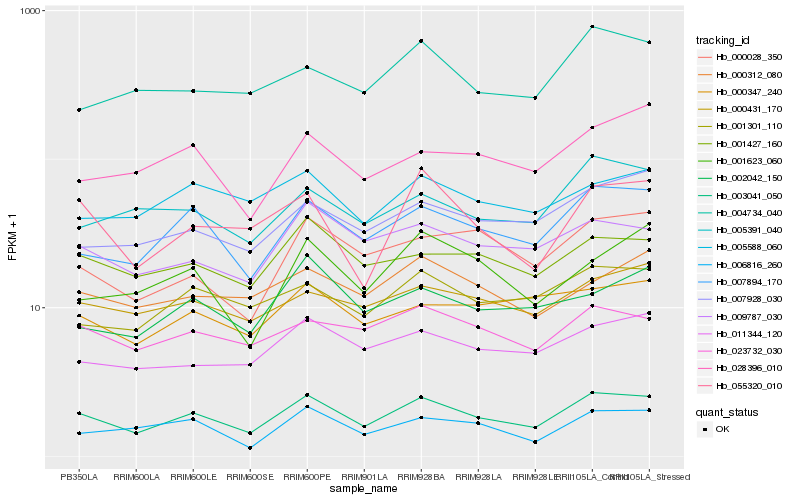
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003041_050 |
0.0 |
- |
- |
- |
| 2 |
Hb_007894_170 |
0.0802249888 |
- |
- |
PREDICTED: UPF0136 membrane protein At2g26240-like [Jatropha curcas] |
| 3 |
Hb_009787_030 |
0.1045092154 |
- |
- |
Prolactin regulatory element-binding protein, putative [Ricinus communis] |
| 4 |
Hb_001427_160 |
0.1069974849 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 5 |
Hb_011344_120 |
0.1076404336 |
- |
- |
PREDICTED: uncharacterized protein LOC105645969 [Jatropha curcas] |
| 6 |
Hb_001301_110 |
0.1104575206 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 22 homolog 1 [Jatropha curcas] |
| 7 |
Hb_055320_010 |
0.1105860998 |
- |
- |
PREDICTED: oligosaccharyltransferase complex subunit ostc [Jatropha curcas] |
| 8 |
Hb_023732_030 |
0.1109485503 |
- |
- |
PREDICTED: proteinaceous RNase P 2-like isoform X2 [Jatropha curcas] |
| 9 |
Hb_000028_350 |
0.1120692565 |
- |
- |
PREDICTED: uncharacterized protein LOC105634642 [Jatropha curcas] |
| 10 |
Hb_000431_170 |
0.1124833483 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein 511 [Jatropha curcas] |
| 11 |
Hb_007928_030 |
0.1126218708 |
- |
- |
PREDICTED: dual specificity phosphatase Cdc25 [Jatropha curcas] |
| 12 |
Hb_006816_260 |
0.1126623502 |
- |
- |
hypothetical protein JCGZ_23244 [Jatropha curcas] |
| 13 |
Hb_005391_040 |
0.1131335342 |
- |
- |
PREDICTED: 1,4-dihydroxy-2-naphthoyl-CoA thioesterase 1 [Jatropha curcas] |
| 14 |
Hb_002042_150 |
0.1171896242 |
- |
- |
PREDICTED: signal peptidase complex subunit 3B [Jatropha curcas] |
| 15 |
Hb_028396_010 |
0.1174961225 |
- |
- |
PREDICTED: probable ATP synthase 24 kDa subunit, mitochondrial [Vitis vinifera] |
| 16 |
Hb_000347_240 |
0.1176209223 |
- |
- |
PREDICTED: proteasome subunit alpha type-6 [Pyrus x bretschneideri] |
| 17 |
Hb_005588_060 |
0.1178490802 |
- |
- |
PREDICTED: succinate dehydrogenase subunit 5, mitochondrial [Jatropha curcas] |
| 18 |
Hb_000312_080 |
0.11800823 |
- |
- |
PREDICTED: transmembrane protein 97-like [Jatropha curcas] |
| 19 |
Hb_001623_060 |
0.1200292479 |
- |
- |
potassium channel beta, putative [Ricinus communis] |
| 20 |
Hb_004734_040 |
0.121139003 |
- |
- |
eukaryotic translation initiation factor 5A isoform I [Hevea brasiliensis] |