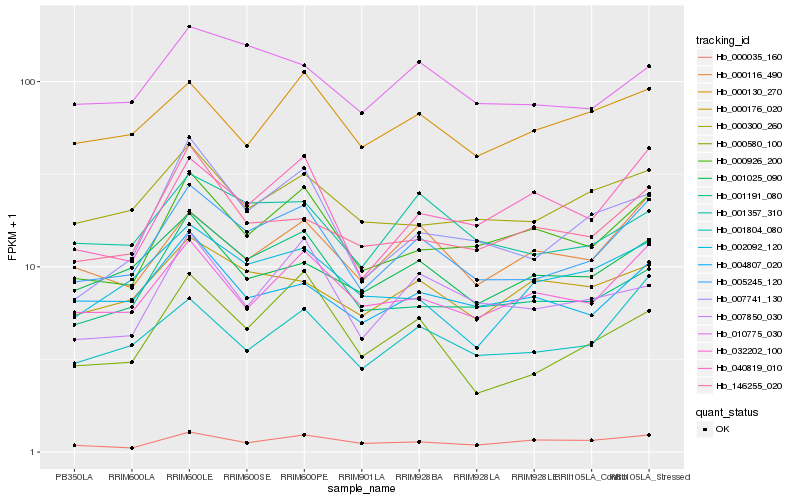
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005245_120 |
0.0 |
- |
- |
XPA-binding protein, putative [Ricinus communis] |
| 2 |
Hb_032202_100 |
0.0909623828 |
desease resistance |
Gene Name: ArsA_ATPase |
arsenical pump-driving atpase, putative [Ricinus communis] |
| 3 |
Hb_000926_200 |
0.0915620992 |
transcription factor |
TF Family: Rcd1-like |
PREDICTED: cell differentiation protein RCD1 homolog isoform X1 [Jatropha curcas] |
| 4 |
Hb_001804_080 |
0.0923444027 |
- |
- |
PREDICTED: aldo-keto reductase family 4 member C9-like [Jatropha curcas] |
| 5 |
Hb_000116_490 |
0.0972857427 |
- |
- |
PREDICTED: WD repeat-containing protein 55 homolog [Jatropha curcas] |
| 6 |
Hb_001357_310 |
0.0974268267 |
- |
- |
PREDICTED: dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex 2, mitochondrial-like [Jatropha curcas] |
| 7 |
Hb_001191_080 |
0.0999014853 |
- |
- |
protein with unknown function [Ricinus communis] |
| 8 |
Hb_007741_130 |
0.10205375 |
- |
- |
hypothetical protein B456_007G089000 [Gossypium raimondii] |
| 9 |
Hb_010775_030 |
0.1031373733 |
- |
- |
PREDICTED: 60S ribosomal protein L4 [Vitis vinifera] |
| 10 |
Hb_040819_010 |
0.1042273598 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 11 |
Hb_000580_100 |
0.1050717036 |
- |
- |
PREDICTED: uncharacterized protein LOC105643133 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000300_260 |
0.1052650357 |
- |
- |
PREDICTED: replication factor C subunit 3 [Jatropha curcas] |
| 13 |
Hb_004807_020 |
0.1061663298 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 14b-like [Jatropha curcas] |
| 14 |
Hb_001025_090 |
0.109411757 |
- |
- |
PREDICTED: FKBP12-interacting protein of 37 kDa [Jatropha curcas] |
| 15 |
Hb_146255_020 |
0.1101450723 |
- |
- |
PREDICTED: chromatin structure-remodeling complex protein BSH [Jatropha curcas] |
| 16 |
Hb_002092_120 |
0.110220395 |
- |
- |
PREDICTED: peptidyl-tRNA hydrolase, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000130_270 |
0.1106899933 |
- |
- |
PREDICTED: malate dehydrogenase, mitochondrial [Jatropha curcas] |
| 18 |
Hb_000176_020 |
0.1120706435 |
- |
- |
PREDICTED: nuclear pore complex protein NUP43 [Jatropha curcas] |
| 19 |
Hb_007850_030 |
0.112269041 |
- |
- |
PREDICTED: uncharacterized protein LOC105640669 [Jatropha curcas] |
| 20 |
Hb_000035_160 |
0.1137688212 |
- |
- |
PREDICTED: formin-like protein 18 [Jatropha curcas] |