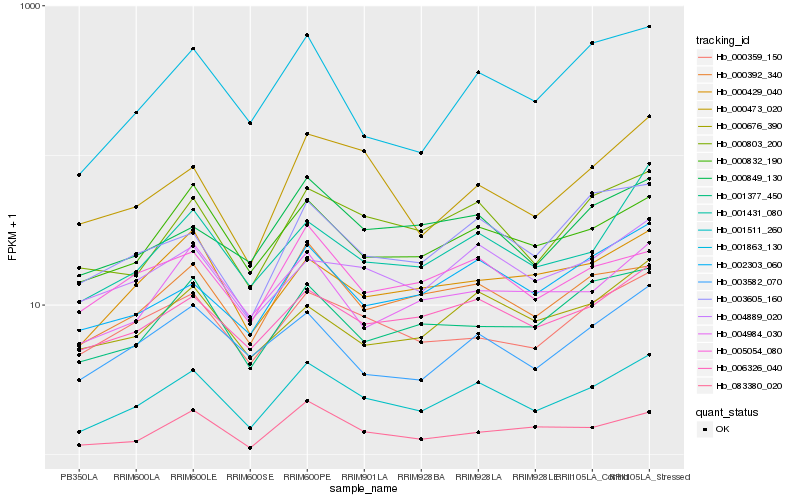
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001511_260 |
0.0 |
- |
- |
tryptophan biosynthesis protein, putative [Ricinus communis] |
| 2 |
Hb_000803_200 |
0.0947271339 |
- |
- |
ubiquitin-conjugating enzyme E2 C, putative [Ricinus communis] |
| 3 |
Hb_006326_040 |
0.1008207977 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000392_340 |
0.1010905675 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 29 [Jatropha curcas] |
| 5 |
Hb_000832_190 |
0.1011263965 |
- |
- |
Rab3 [Hevea brasiliensis] |
| 6 |
Hb_000359_150 |
0.1094738298 |
- |
- |
PREDICTED: protein Mpv17-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_002303_060 |
0.1128058134 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000429_040 |
0.1144482471 |
- |
- |
PREDICTED: chaperone protein dnaJ 6 [Vitis vinifera] |
| 9 |
Hb_003582_070 |
0.1145602353 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001377_450 |
0.11588316 |
- |
- |
PREDICTED: pyridoxine/pyridoxamine 5'-phosphate oxidase 2 isoform X1 [Jatropha curcas] |
| 11 |
Hb_004889_020 |
0.1160607869 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 20 homolog 2-like [Jatropha curcas] |
| 12 |
Hb_001431_080 |
0.1166869445 |
- |
- |
40S ribosomal protein S11, putative [Ricinus communis] |
| 13 |
Hb_000849_130 |
0.1178779454 |
- |
- |
PREDICTED: adrenodoxin-like protein, mitochondrial [Nelumbo nucifera] |
| 14 |
Hb_001863_130 |
0.1186649418 |
- |
- |
hypothetical protein ZEAMMB73_433257, partial [Zea mays] |
| 15 |
Hb_003605_160 |
0.11900141 |
- |
- |
PREDICTED: uncharacterized protein LOC105135144 [Populus euphratica] |
| 16 |
Hb_005054_080 |
0.1191598597 |
transcription factor |
TF Family: LIM |
Pollen-specific protein SF3, putative [Ricinus communis] |
| 17 |
Hb_000473_020 |
0.1197571951 |
- |
- |
expressed protein, putative [Ricinus communis] |
| 18 |
Hb_004984_030 |
0.1198937636 |
- |
- |
protein with unknown function [Ricinus communis] |
| 19 |
Hb_000676_390 |
0.1204626981 |
- |
- |
PREDICTED: protein FMP32, mitochondrial [Jatropha curcas] |
| 20 |
Hb_083380_020 |
0.1204922866 |
- |
- |
PREDICTED: LAG1 longevity assurance homolog 2 [Jatropha curcas] |