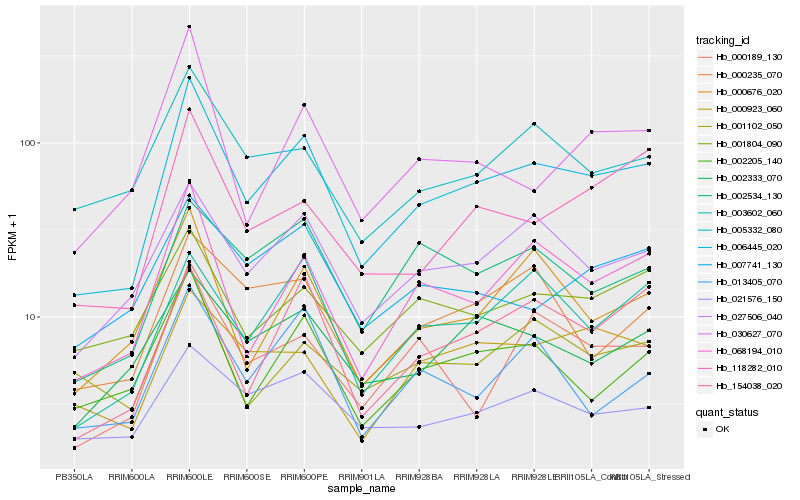
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006445_020 |
0.0 |
- |
- |
hypothetical protein L484_026741 [Morus notabilis] |
| 2 |
Hb_068194_010 |
0.1117900903 |
- |
- |
hypothetical protein POPTR_0010s23740g [Populus trichocarpa] |
| 3 |
Hb_027506_040 |
0.1209850567 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit beta-3, chloroplastic [Jatropha curcas] |
| 4 |
Hb_002205_140 |
0.1294211984 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor RF2b [Jatropha curcas] |
| 5 |
Hb_000676_020 |
0.130618961 |
- |
- |
PREDICTED: uncharacterized protein LOC105645386 [Jatropha curcas] |
| 6 |
Hb_003602_060 |
0.1394088975 |
- |
- |
PREDICTED: phosphoglycerate mutase-like protein 1 [Jatropha curcas] |
| 7 |
Hb_118282_010 |
0.1407094725 |
- |
- |
PREDICTED: uncharacterized protein LOC105108358 [Populus euphratica] |
| 8 |
Hb_154038_020 |
0.1411363465 |
- |
- |
hypothetical protein JCGZ_05648 [Jatropha curcas] |
| 9 |
Hb_005332_080 |
0.1415449763 |
- |
- |
PREDICTED: succinate dehydrogenase [ubiquinone] iron-sulfur subunit 2, mitochondrial [Jatropha curcas] |
| 10 |
Hb_002333_070 |
0.143181725 |
- |
- |
PREDICTED: ACT domain-containing protein ACR4-like [Jatropha curcas] |
| 11 |
Hb_021576_150 |
0.1432797567 |
- |
- |
PREDICTED: formin-like protein 18 [Jatropha curcas] |
| 12 |
Hb_030627_070 |
0.1437156358 |
- |
- |
histone H4 [Zea mays] |
| 13 |
Hb_000189_130 |
0.1470247074 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATA, chloroplastic [Jatropha curcas] |
| 14 |
Hb_001804_090 |
0.147532021 |
- |
- |
PREDICTED: uracil phosphoribosyltransferase isoform X1 [Jatropha curcas] |
| 15 |
Hb_002534_130 |
0.1475661576 |
- |
- |
membrane associated ring finger 1,8, putative [Ricinus communis] |
| 16 |
Hb_013405_070 |
0.1477139737 |
- |
- |
ferric-chelate reductase, putative [Ricinus communis] |
| 17 |
Hb_000235_070 |
0.147791481 |
- |
- |
PREDICTED: VIN3-like protein 2 [Jatropha curcas] |
| 18 |
Hb_007741_130 |
0.1486962473 |
- |
- |
hypothetical protein B456_007G089000 [Gossypium raimondii] |
| 19 |
Hb_000923_060 |
0.1489521652 |
- |
- |
PREDICTED: uncharacterized protein LOC105628271 [Jatropha curcas] |
| 20 |
Hb_001102_050 |
0.149113853 |
- |
- |
PREDICTED: fatty-acid-binding protein 3 [Jatropha curcas] |