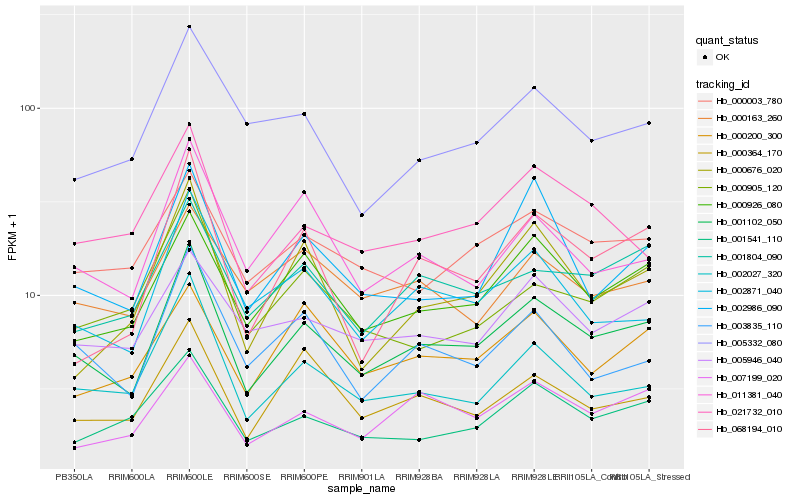
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001102_050 |
0.0 |
- |
- |
PREDICTED: fatty-acid-binding protein 3 [Jatropha curcas] |
| 2 |
Hb_000926_080 |
0.1042589799 |
- |
- |
PREDICTED: paraspeckle component 1 [Jatropha curcas] |
| 3 |
Hb_002027_320 |
0.1047330685 |
- |
- |
PREDICTED: RNA-binding protein BRN1 [Jatropha curcas] |
| 4 |
Hb_002871_040 |
0.108937174 |
- |
- |
PREDICTED: enoyl-[acyl-carrier-protein] reductase [NADH], chloroplastic-like [Populus euphratica] |
| 5 |
Hb_001804_090 |
0.1105569446 |
- |
- |
PREDICTED: uracil phosphoribosyltransferase isoform X1 [Jatropha curcas] |
| 6 |
Hb_003835_110 |
0.1129491335 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 11 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000003_780 |
0.1163143751 |
- |
- |
hexokinase [Manihot esculenta] |
| 8 |
Hb_000364_170 |
0.1174404118 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase CLF [Jatropha curcas] |
| 9 |
Hb_001541_110 |
0.1179045121 |
- |
- |
PREDICTED: sufE-like protein 2, chloroplastic isoform X2 [Jatropha curcas] |
| 10 |
Hb_011381_040 |
0.1189383556 |
- |
- |
PREDICTED: uncharacterized protein At1g32220, chloroplastic [Jatropha curcas] |
| 11 |
Hb_007199_020 |
0.1198921393 |
- |
- |
hypothetical protein CICLE_v10028570mg [Citrus clementina] |
| 12 |
Hb_005946_040 |
0.1198955352 |
- |
- |
PREDICTED: probable protein phosphatase 2C 22 isoform X2 [Jatropha curcas] |
| 13 |
Hb_021732_010 |
0.1208468455 |
- |
- |
biotin carboxylase [Vernicia fordii] |
| 14 |
Hb_068194_010 |
0.1226018651 |
- |
- |
hypothetical protein POPTR_0010s23740g [Populus trichocarpa] |
| 15 |
Hb_002986_090 |
0.123431873 |
- |
- |
PREDICTED: methionine aminopeptidase 1D, chloroplastic/mitochondrial [Jatropha curcas] |
| 16 |
Hb_005332_080 |
0.1239993023 |
- |
- |
PREDICTED: succinate dehydrogenase [ubiquinone] iron-sulfur subunit 2, mitochondrial [Jatropha curcas] |
| 17 |
Hb_000905_120 |
0.1247119873 |
- |
- |
PREDICTED: uncharacterized protein LOC105628677 [Jatropha curcas] |
| 18 |
Hb_000676_020 |
0.1291703248 |
- |
- |
PREDICTED: uncharacterized protein LOC105645386 [Jatropha curcas] |
| 19 |
Hb_000163_260 |
0.1292134896 |
- |
- |
PREDICTED: uncharacterized protein LOC105642518 [Jatropha curcas] |
| 20 |
Hb_000200_300 |
0.130983358 |
- |
- |
PREDICTED: uncharacterized protein LOC105636926 [Jatropha curcas] |