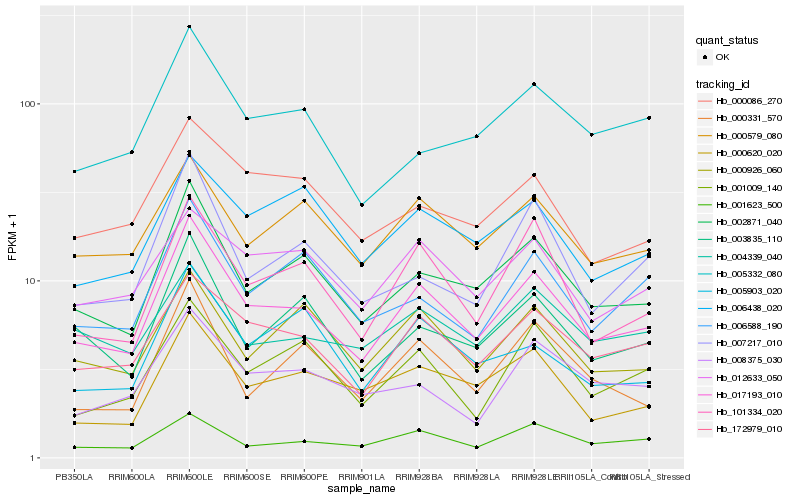
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_017193_010 |
0.0 |
- |
- |
PREDICTED: lactation elevated protein 1 isoform X1 [Jatropha curcas] |
| 2 |
Hb_002871_040 |
0.0769086226 |
- |
- |
PREDICTED: enoyl-[acyl-carrier-protein] reductase [NADH], chloroplastic-like [Populus euphratica] |
| 3 |
Hb_172979_010 |
0.0863872226 |
- |
- |
PREDICTED: protein disulfide-isomerase SCO2 [Jatropha curcas] |
| 4 |
Hb_101334_020 |
0.0956076754 |
- |
- |
PREDICTED: uncharacterized WD repeat-containing protein C2A9.03-like [Jatropha curcas] |
| 5 |
Hb_001623_500 |
0.1056948619 |
- |
- |
PREDICTED: uncharacterized protein LOC105638487 [Jatropha curcas] |
| 6 |
Hb_003835_110 |
0.1084034451 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 11 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000579_080 |
0.1084509874 |
- |
- |
PREDICTED: 26S protease regulatory subunit S10B homolog B [Jatropha curcas] |
| 8 |
Hb_000620_020 |
0.1128039619 |
- |
- |
PREDICTED: uncharacterized protein LOC105649917 [Jatropha curcas] |
| 9 |
Hb_000926_060 |
0.1159932398 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000086_270 |
0.1168352388 |
- |
- |
hypothetical protein POPTR_0002s03730g [Populus trichocarpa] |
| 11 |
Hb_012633_050 |
0.116836296 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 12 |
Hb_007217_010 |
0.1182902494 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase, chloroplastic [Jatropha curcas] |
| 13 |
Hb_001009_140 |
0.1194702121 |
- |
- |
hypothetical protein JCGZ_11337 [Jatropha curcas] |
| 14 |
Hb_006588_190 |
0.1197961306 |
- |
- |
PREDICTED: ataxin-3 homolog [Jatropha curcas] |
| 15 |
Hb_005332_080 |
0.1202921678 |
- |
- |
PREDICTED: succinate dehydrogenase [ubiquinone] iron-sulfur subunit 2, mitochondrial [Jatropha curcas] |
| 16 |
Hb_000331_570 |
0.1223959382 |
- |
- |
PREDICTED: DNA-(apurinic or apyrimidinic site) lyase [Jatropha curcas] |
| 17 |
Hb_008375_030 |
0.1232671217 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 4 isoform X1 [Populus euphratica] |
| 18 |
Hb_006438_020 |
0.1234325525 |
- |
- |
PREDICTED: lysine--tRNA ligase-like [Populus euphratica] |
| 19 |
Hb_004339_040 |
0.1248246117 |
- |
- |
hypothetical protein POPTR_0162s00290g, partial [Populus trichocarpa] |
| 20 |
Hb_005903_020 |
0.1256136564 |
- |
- |
12-oxophytodienoate reductase 2 [Theobroma cacao] |