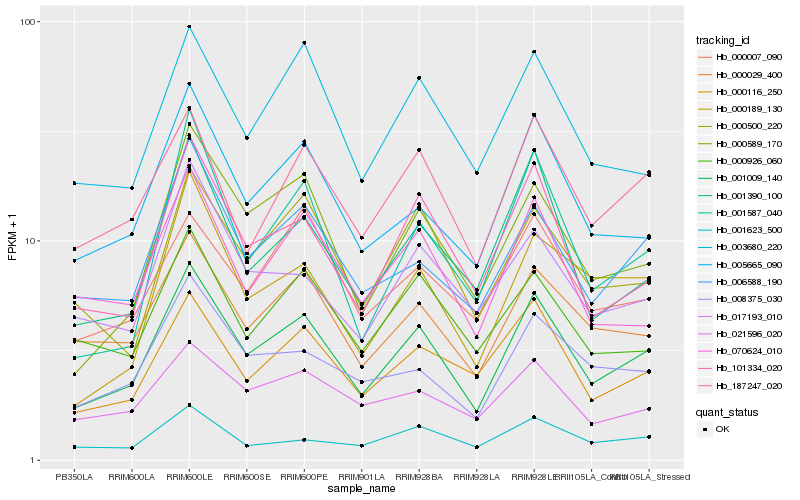
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001009_140 |
0.0 |
- |
- |
hypothetical protein JCGZ_11337 [Jatropha curcas] |
| 2 |
Hb_005665_090 |
0.0940004327 |
- |
- |
PREDICTED: 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase isoform X2 [Jatropha curcas] |
| 3 |
Hb_101334_020 |
0.09402375 |
- |
- |
PREDICTED: uncharacterized WD repeat-containing protein C2A9.03-like [Jatropha curcas] |
| 4 |
Hb_000589_170 |
0.0964193668 |
- |
- |
PREDICTED: telomere repeat-binding protein 4 isoform X3 [Jatropha curcas] |
| 5 |
Hb_000029_400 |
0.0974856415 |
- |
- |
PREDICTED: probable 1-acyl-sn-glycerol-3-phosphate acyltransferase 5 isoform X1 [Jatropha curcas] |
| 6 |
Hb_006588_190 |
0.0990613081 |
- |
- |
PREDICTED: ataxin-3 homolog [Jatropha curcas] |
| 7 |
Hb_000116_250 |
0.099267552 |
- |
- |
PREDICTED: DNA mismatch repair protein MLH3 [Jatropha curcas] |
| 8 |
Hb_187247_020 |
0.1021708551 |
- |
- |
PREDICTED: uncharacterized protein LOC101220469 [Cucumis sativus] |
| 9 |
Hb_001390_100 |
0.1033903655 |
- |
- |
PREDICTED: LL-diaminopimelate aminotransferase, chloroplastic [Jatropha curcas] |
| 10 |
Hb_003680_220 |
0.1083870018 |
- |
- |
hypothetical protein B456_002G243700 [Gossypium raimondii] |
| 11 |
Hb_008375_030 |
0.1089390038 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 4 isoform X1 [Populus euphratica] |
| 12 |
Hb_070624_010 |
0.1102194614 |
- |
- |
PREDICTED: reticulocalbin-2 [Jatropha curcas] |
| 13 |
Hb_000500_220 |
0.1103752181 |
- |
- |
PREDICTED: intersectin-1 [Jatropha curcas] |
| 14 |
Hb_021596_020 |
0.1110431886 |
- |
- |
hypothetical protein JCGZ_02034 [Jatropha curcas] |
| 15 |
Hb_001587_040 |
0.1127217858 |
- |
- |
PREDICTED: soluble inorganic pyrophosphatase 6, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000926_060 |
0.1159497821 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000189_130 |
0.1160111062 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATA, chloroplastic [Jatropha curcas] |
| 18 |
Hb_017193_010 |
0.1194702121 |
- |
- |
PREDICTED: lactation elevated protein 1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001623_500 |
0.1195950803 |
- |
- |
PREDICTED: uncharacterized protein LOC105638487 [Jatropha curcas] |
| 20 |
Hb_000007_090 |
0.1197105499 |
- |
- |
PREDICTED: LAG1 longevity assurance homolog 3 [Jatropha curcas] |