| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
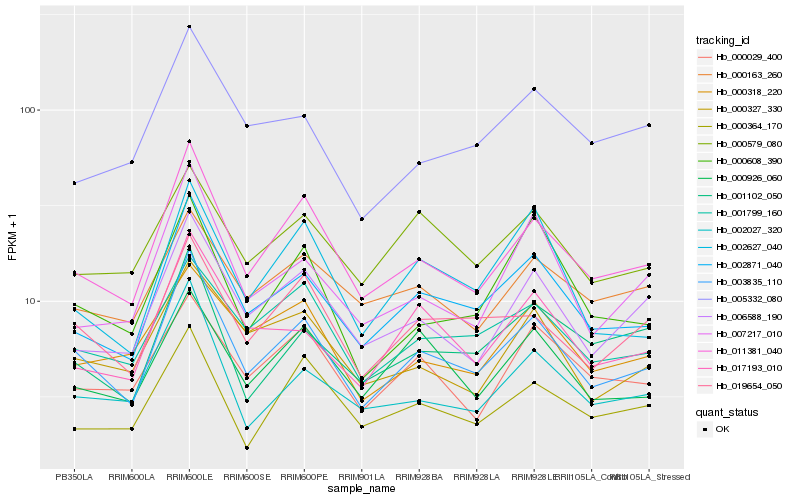
Hb_003835_110 |
0.0 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 11 isoform X1 [Jatropha curcas] |
| 2 |
Hb_011381_040 |
0.0650160054 |
- |
- |
PREDICTED: uncharacterized protein At1g32220, chloroplastic [Jatropha curcas] |
| 3 |
Hb_002871_040 |
0.0670144906 |
- |
- |
PREDICTED: enoyl-[acyl-carrier-protein] reductase [NADH], chloroplastic-like [Populus euphratica] |
| 4 |
Hb_019654_050 |
0.0985517118 |
- |
- |
PREDICTED: prolyl endopeptidase [Jatropha curcas] |
| 5 |
Hb_002027_320 |
0.1016169398 |
- |
- |
PREDICTED: RNA-binding protein BRN1 [Jatropha curcas] |
| 6 |
Hb_006588_190 |
0.1051287808 |
- |
- |
PREDICTED: ataxin-3 homolog [Jatropha curcas] |
| 7 |
Hb_017193_010 |
0.1084034451 |
- |
- |
PREDICTED: lactation elevated protein 1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000327_330 |
0.1103655592 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001799_160 |
0.110879459 |
- |
- |
PREDICTED: protein VASCULAR ASSOCIATED DEATH 1, chloroplastic isoform X2 [Jatropha curcas] |
| 10 |
Hb_001102_050 |
0.1129491335 |
- |
- |
PREDICTED: fatty-acid-binding protein 3 [Jatropha curcas] |
| 11 |
Hb_002627_040 |
0.1155641215 |
- |
- |
prolyl 4-hydroxylase alpha subunit, putative [Ricinus communis] |
| 12 |
Hb_007217_010 |
0.1156016721 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000608_390 |
0.1156335832 |
- |
- |
hypothetical protein B456_010G140700 [Gossypium raimondii] |
| 14 |
Hb_000926_060 |
0.1160471663 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000364_170 |
0.1193646351 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase CLF [Jatropha curcas] |
| 16 |
Hb_000029_400 |
0.1205976995 |
- |
- |
PREDICTED: probable 1-acyl-sn-glycerol-3-phosphate acyltransferase 5 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000579_080 |
0.1223232812 |
- |
- |
PREDICTED: 26S protease regulatory subunit S10B homolog B [Jatropha curcas] |
| 18 |
Hb_000318_220 |
0.1241428802 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_005332_080 |
0.1258140044 |
- |
- |
PREDICTED: succinate dehydrogenase [ubiquinone] iron-sulfur subunit 2, mitochondrial [Jatropha curcas] |
| 20 |
Hb_000163_260 |
0.1260071029 |
- |
- |
PREDICTED: uncharacterized protein LOC105642518 [Jatropha curcas] |