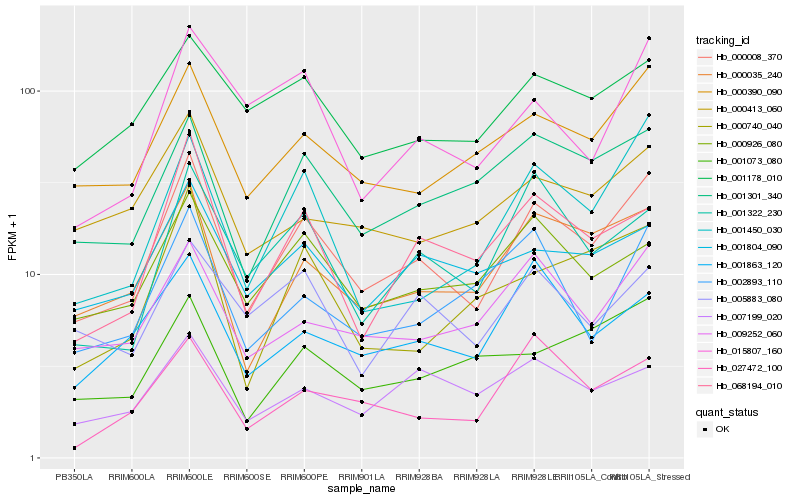
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000008_370 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105628095 [Jatropha curcas] |
| 2 |
Hb_000035_240 |
0.1129925684 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000390_090 |
0.1147642333 |
- |
- |
PREDICTED: uncharacterized protein LOC105632476 [Jatropha curcas] |
| 4 |
Hb_009252_060 |
0.1272117716 |
- |
- |
PREDICTED: uncharacterized protein LOC105640395 [Jatropha curcas] |
| 5 |
Hb_001301_340 |
0.1288122757 |
- |
- |
endoribonuclease L-PSP family protein [Populus trichocarpa] |
| 6 |
Hb_001450_030 |
0.1298594283 |
- |
- |
PREDICTED: uncharacterized protein LOC105650827 [Jatropha curcas] |
| 7 |
Hb_068194_010 |
0.1322509054 |
- |
- |
hypothetical protein POPTR_0010s23740g [Populus trichocarpa] |
| 8 |
Hb_000926_080 |
0.1363432899 |
- |
- |
PREDICTED: paraspeckle component 1 [Jatropha curcas] |
| 9 |
Hb_001322_230 |
0.1372918859 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 4, chloroplastic [Populus euphratica] |
| 10 |
Hb_001804_090 |
0.1375302565 |
- |
- |
PREDICTED: uracil phosphoribosyltransferase isoform X1 [Jatropha curcas] |
| 11 |
Hb_001178_010 |
0.1413931052 |
- |
- |
PREDICTED: uncharacterized protein LOC105629461 [Jatropha curcas] |
| 12 |
Hb_001863_120 |
0.1427825715 |
- |
- |
PREDICTED: serine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 13 |
Hb_015807_160 |
0.1434383905 |
- |
- |
PREDICTED: probable histone H2A.1 [Jatropha curcas] |
| 14 |
Hb_001073_080 |
0.1453135396 |
- |
- |
PREDICTED: protein root UVB sensitive 2, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000740_040 |
0.1465483717 |
- |
- |
PREDICTED: uncharacterized protein LOC105126082 [Populus euphratica] |
| 16 |
Hb_002893_110 |
0.1472266103 |
transcription factor |
TF Family: DBP |
PREDICTED: probable protein phosphatase 2C 22 isoform X2 [Jatropha curcas] |
| 17 |
Hb_027472_100 |
0.1473252697 |
- |
- |
PREDICTED: uncharacterized protein LOC105646173 [Jatropha curcas] |
| 18 |
Hb_007199_020 |
0.1473441511 |
- |
- |
hypothetical protein CICLE_v10028570mg [Citrus clementina] |
| 19 |
Hb_000413_060 |
0.147349814 |
- |
- |
PREDICTED: DAG protein, chloroplastic [Populus euphratica] |
| 20 |
Hb_005883_080 |
0.1477226165 |
- |
- |
type 2A protein phosphatase-2 [Populus trichocarpa] |