| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
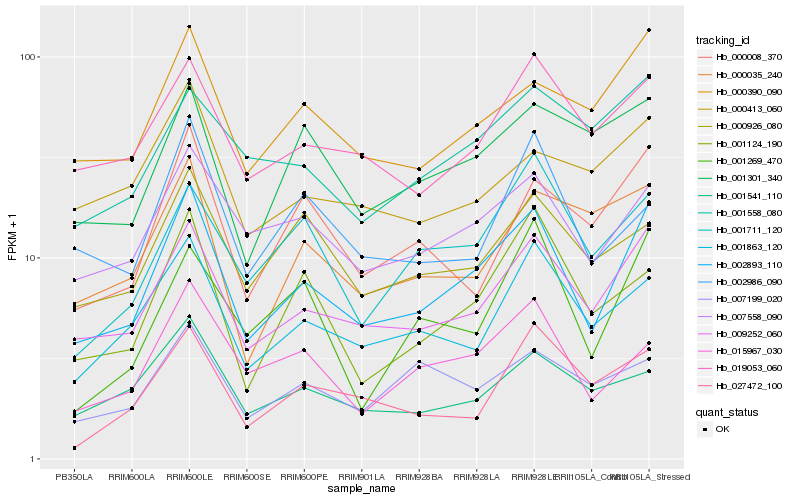
Hb_002893_110 |
0.0 |
transcription factor |
TF Family: DBP |
PREDICTED: probable protein phosphatase 2C 22 isoform X2 [Jatropha curcas] |
| 2 |
Hb_009252_060 |
0.0844769361 |
- |
- |
PREDICTED: uncharacterized protein LOC105640395 [Jatropha curcas] |
| 3 |
Hb_007558_090 |
0.1260659162 |
- |
- |
PREDICTED: serine/threonine-protein kinase STN8, chloroplastic isoform X1 [Jatropha curcas] |
| 4 |
Hb_000390_090 |
0.1282636974 |
- |
- |
PREDICTED: uncharacterized protein LOC105632476 [Jatropha curcas] |
| 5 |
Hb_001863_120 |
0.131886427 |
- |
- |
PREDICTED: serine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 6 |
Hb_019053_060 |
0.1338212952 |
- |
- |
PREDICTED: uncharacterized protein LOC105632672 [Jatropha curcas] |
| 7 |
Hb_015967_030 |
0.1343183028 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription repressor VAL1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000008_370 |
0.1472266103 |
- |
- |
PREDICTED: uncharacterized protein LOC105628095 [Jatropha curcas] |
| 9 |
Hb_001124_190 |
0.1475046999 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000926_080 |
0.1495390536 |
- |
- |
PREDICTED: paraspeckle component 1 [Jatropha curcas] |
| 11 |
Hb_001541_110 |
0.1511924778 |
- |
- |
PREDICTED: sufE-like protein 2, chloroplastic isoform X2 [Jatropha curcas] |
| 12 |
Hb_001711_120 |
0.1522149899 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 4, chloroplastic [Jatropha curcas] |
| 13 |
Hb_001301_340 |
0.1536634205 |
- |
- |
endoribonuclease L-PSP family protein [Populus trichocarpa] |
| 14 |
Hb_000035_240 |
0.1571708796 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000413_060 |
0.1572922688 |
- |
- |
PREDICTED: DAG protein, chloroplastic [Populus euphratica] |
| 16 |
Hb_001558_080 |
0.1574948352 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1D, chloroplastic isoform X1 [Jatropha curcas] |
| 17 |
Hb_002986_090 |
0.1581550311 |
- |
- |
PREDICTED: methionine aminopeptidase 1D, chloroplastic/mitochondrial [Jatropha curcas] |
| 18 |
Hb_001269_470 |
0.1582497467 |
- |
- |
PREDICTED: uncharacterized protein LOC105630313 [Jatropha curcas] |
| 19 |
Hb_027472_100 |
0.1608224112 |
- |
- |
PREDICTED: uncharacterized protein LOC105646173 [Jatropha curcas] |
| 20 |
Hb_007199_020 |
0.1609638355 |
- |
- |
hypothetical protein CICLE_v10028570mg [Citrus clementina] |