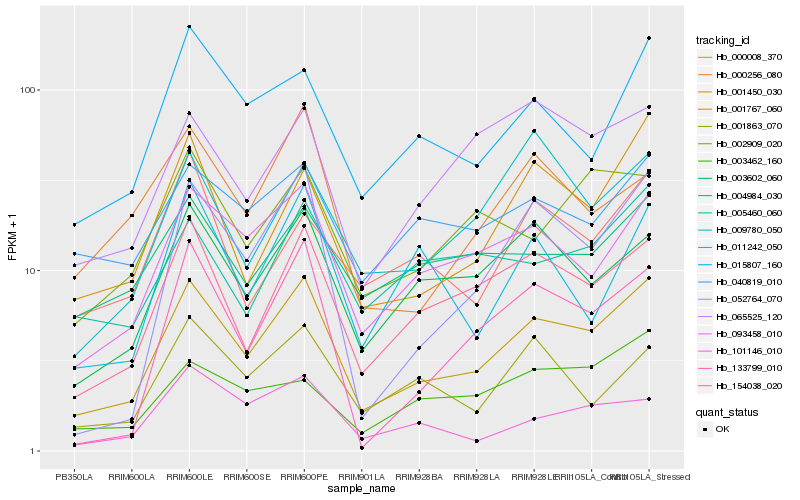
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001767_060 |
0.0 |
- |
- |
PREDICTED: sulfate transporter 4.1, chloroplastic-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_093458_010 |
0.1136320791 |
- |
- |
hypothetical protein CISIN_1g0277592mg [Citrus sinensis] |
| 3 |
Hb_154038_020 |
0.1136762754 |
- |
- |
hypothetical protein JCGZ_05648 [Jatropha curcas] |
| 4 |
Hb_003602_060 |
0.1323936391 |
- |
- |
PREDICTED: phosphoglycerate mutase-like protein 1 [Jatropha curcas] |
| 5 |
Hb_001450_030 |
0.1331786408 |
- |
- |
PREDICTED: uncharacterized protein LOC105650827 [Jatropha curcas] |
| 6 |
Hb_015807_160 |
0.1378068275 |
- |
- |
PREDICTED: probable histone H2A.1 [Jatropha curcas] |
| 7 |
Hb_052764_070 |
0.1397319106 |
- |
- |
PREDICTED: uncharacterized protein LOC105649473 [Jatropha curcas] |
| 8 |
Hb_133799_010 |
0.1555811263 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_004984_030 |
0.1562674429 |
- |
- |
protein with unknown function [Ricinus communis] |
| 10 |
Hb_040819_010 |
0.1562786141 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 11 |
Hb_065525_120 |
0.1566021898 |
- |
- |
dtdp-glucose 4-6-dehydratase, putative [Ricinus communis] |
| 12 |
Hb_001863_070 |
0.159510964 |
- |
- |
Queuine tRNA-ribosyltransferase [Theobroma cacao] |
| 13 |
Hb_002909_020 |
0.1615553366 |
- |
- |
PREDICTED: uncharacterized protein LOC105632476 [Jatropha curcas] |
| 14 |
Hb_009780_050 |
0.1621826604 |
- |
- |
PREDICTED: glutathione S-transferase DHAR3, chloroplastic [Jatropha curcas] |
| 15 |
Hb_005460_060 |
0.1627986474 |
- |
- |
PREDICTED: proteasome assembly chaperone 2 [Jatropha curcas] |
| 16 |
Hb_101146_010 |
0.1628531777 |
- |
- |
PREDICTED: peregrin-like [Jatropha curcas] |
| 17 |
Hb_011242_050 |
0.1644388126 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2-23 kDa [Jatropha curcas] |
| 18 |
Hb_003462_160 |
0.1647021196 |
transcription factor |
TF Family: GNAT |
predicted protein [Arabidopsis lyrata subsp. lyrata] |
| 19 |
Hb_000008_370 |
0.1653142912 |
- |
- |
PREDICTED: uncharacterized protein LOC105628095 [Jatropha curcas] |
| 20 |
Hb_000256_080 |
0.1654351411 |
- |
- |
PREDICTED: deSI-like protein At4g17486 [Jatropha curcas] |