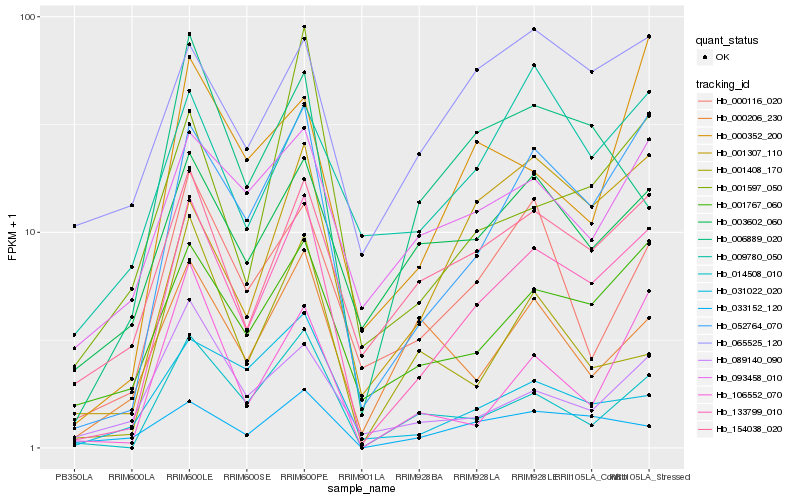
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_133799_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_052764_070 |
0.0926084109 |
- |
- |
PREDICTED: uncharacterized protein LOC105649473 [Jatropha curcas] |
| 3 |
Hb_001767_060 |
0.1555811263 |
- |
- |
PREDICTED: sulfate transporter 4.1, chloroplastic-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_154038_020 |
0.1647279755 |
- |
- |
hypothetical protein JCGZ_05648 [Jatropha curcas] |
| 5 |
Hb_014508_010 |
0.1666977382 |
- |
- |
PREDICTED: uncharacterized protein LOC105629998 [Jatropha curcas] |
| 6 |
Hb_001307_110 |
0.1767079445 |
- |
- |
PREDICTED: uncharacterized protein LOC105644507 [Jatropha curcas] |
| 7 |
Hb_033152_120 |
0.1890376174 |
- |
- |
PREDICTED: la-related protein 6C [Jatropha curcas] |
| 8 |
Hb_003602_060 |
0.1922407446 |
- |
- |
PREDICTED: phosphoglycerate mutase-like protein 1 [Jatropha curcas] |
| 9 |
Hb_000116_020 |
0.1940431465 |
- |
- |
PREDICTED: serine acetyltransferase 2-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_106552_070 |
0.1980501032 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_093458_010 |
0.2013020659 |
- |
- |
hypothetical protein CISIN_1g0277592mg [Citrus sinensis] |
| 12 |
Hb_006889_020 |
0.2045781459 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 13 |
Hb_089140_090 |
0.205894035 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360-like isoform X3 [Beta vulgaris subsp. vulgaris] |
| 14 |
Hb_065525_120 |
0.2100741521 |
- |
- |
dtdp-glucose 4-6-dehydratase, putative [Ricinus communis] |
| 15 |
Hb_000352_200 |
0.2104801522 |
- |
- |
PREDICTED: OTU domain-containing protein DDB_G0284757 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000206_230 |
0.2125934814 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_009780_050 |
0.2135710834 |
- |
- |
PREDICTED: glutathione S-transferase DHAR3, chloroplastic [Jatropha curcas] |
| 18 |
Hb_031022_020 |
0.2137164971 |
- |
- |
- |
| 19 |
Hb_001597_050 |
0.2142906275 |
- |
- |
- |
| 20 |
Hb_001408_170 |
0.2144631041 |
- |
- |
PREDICTED: phosphoglycolate phosphatase 2 [Jatropha curcas] |