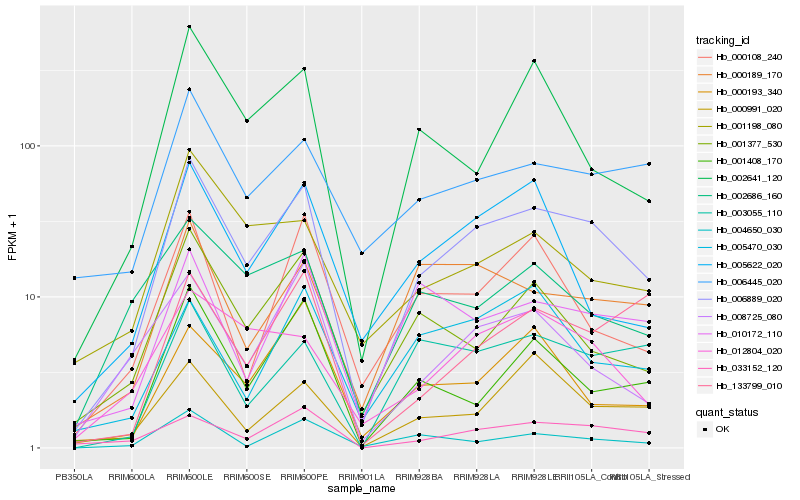
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006889_020 |
0.0 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 2 |
Hb_033152_120 |
0.1660118315 |
- |
- |
PREDICTED: la-related protein 6C [Jatropha curcas] |
| 3 |
Hb_006445_020 |
0.1767381255 |
- |
- |
hypothetical protein L484_026741 [Morus notabilis] |
| 4 |
Hb_001377_530 |
0.1792282399 |
- |
- |
NC domain-containing family protein [Populus trichocarpa] |
| 5 |
Hb_012804_020 |
0.1856867338 |
- |
- |
PREDICTED: abscisic acid 8'-hydroxylase 4-like [Jatropha curcas] |
| 6 |
Hb_000189_170 |
0.186385086 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 7 |
Hb_001408_170 |
0.1870915431 |
- |
- |
PREDICTED: phosphoglycolate phosphatase 2 [Jatropha curcas] |
| 8 |
Hb_010172_110 |
0.189616312 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 9 |
Hb_002641_120 |
0.1921628864 |
- |
- |
PREDICTED: calmodulin [Jatropha curcas] |
| 10 |
Hb_005622_020 |
0.1939775183 |
- |
- |
PREDICTED: DCC family protein At1g52590, chloroplastic [Jatropha curcas] |
| 11 |
Hb_008725_080 |
0.1953470744 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 7 [Jatropha curcas] |
| 12 |
Hb_000991_020 |
0.1981453291 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 13 |
Hb_000108_240 |
0.1985257392 |
- |
- |
hypothetical protein Csa_1G600150 [Cucumis sativus] |
| 14 |
Hb_003055_110 |
0.1993828427 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002686_160 |
0.200187789 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 12-like [Populus euphratica] |
| 16 |
Hb_001198_080 |
0.2008854184 |
- |
- |
PREDICTED: uncharacterized protein LOC105633342 isoform X2 [Jatropha curcas] |
| 17 |
Hb_004650_030 |
0.2010676185 |
- |
- |
PREDICTED: uncharacterized protein LOC105638888 isoform X1 [Jatropha curcas] |
| 18 |
Hb_133799_010 |
0.2045781459 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_005470_030 |
0.2054262124 |
- |
- |
PREDICTED: probable galactinol--sucrose galactosyltransferase 2 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000193_340 |
0.2060695658 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 isoform X1 [Jatropha curcas] |