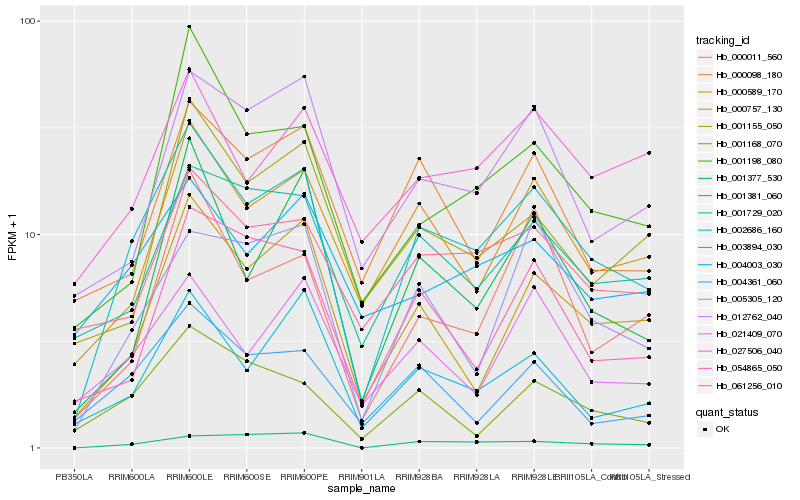
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002686_160 |
0.0 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 12-like [Populus euphratica] |
| 2 |
Hb_000589_170 |
0.143107455 |
- |
- |
PREDICTED: telomere repeat-binding protein 4 isoform X3 [Jatropha curcas] |
| 3 |
Hb_000757_130 |
0.1488003168 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001729_020 |
0.1566852366 |
transcription factor |
TF Family: C3H |
zinc finger protein, putative [Ricinus communis] |
| 5 |
Hb_001377_530 |
0.1577958495 |
- |
- |
NC domain-containing family protein [Populus trichocarpa] |
| 6 |
Hb_003894_030 |
0.1603498733 |
- |
- |
plant sec1, putative [Ricinus communis] |
| 7 |
Hb_005305_120 |
0.1608417294 |
- |
- |
PREDICTED: calcium-dependent protein kinase 8-like [Jatropha curcas] |
| 8 |
Hb_012762_040 |
0.1609563402 |
- |
- |
big map kinase/bmk, putative [Ricinus communis] |
| 9 |
Hb_061256_010 |
0.1616780766 |
- |
- |
PREDICTED: uncharacterized protein LOC105635220 isoform X2 [Jatropha curcas] |
| 10 |
Hb_021409_070 |
0.1653357733 |
- |
- |
PREDICTED: transcription factor bHLH157 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000098_180 |
0.1657080696 |
- |
- |
PREDICTED: uncharacterized protein LOC105633342 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000011_560 |
0.1668698609 |
- |
- |
PREDICTED: phosphoinositide phosphatase SAC2 isoform X1 [Jatropha curcas] |
| 13 |
Hb_004361_060 |
0.1669238321 |
- |
- |
PREDICTED: uncharacterized protein LOC105636705 [Jatropha curcas] |
| 14 |
Hb_054865_050 |
0.1671969932 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 15 |
Hb_001168_070 |
0.1680944103 |
- |
- |
hypothetical protein JCGZ_00277 [Jatropha curcas] |
| 16 |
Hb_004003_030 |
0.1699850577 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001198_080 |
0.1710919764 |
- |
- |
PREDICTED: uncharacterized protein LOC105633342 isoform X2 [Jatropha curcas] |
| 18 |
Hb_001381_060 |
0.1712930245 |
- |
- |
monoxygenase, putative [Ricinus communis] |
| 19 |
Hb_027506_040 |
0.1722063955 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit beta-3, chloroplastic [Jatropha curcas] |
| 20 |
Hb_001155_050 |
0.1743601837 |
- |
- |
PREDICTED: KH domain-containing protein SPIN1-like [Jatropha curcas] |