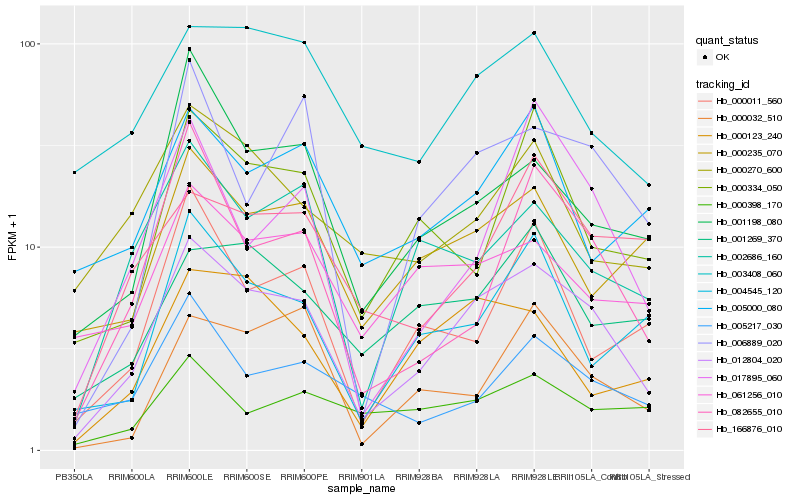
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012804_020 |
0.0 |
- |
- |
PREDICTED: abscisic acid 8'-hydroxylase 4-like [Jatropha curcas] |
| 2 |
Hb_002686_160 |
0.1769562793 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 12-like [Populus euphratica] |
| 3 |
Hb_001198_080 |
0.1856779467 |
- |
- |
PREDICTED: uncharacterized protein LOC105633342 isoform X2 [Jatropha curcas] |
| 4 |
Hb_006889_020 |
0.1856867338 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 5 |
Hb_003408_060 |
0.1865526889 |
transcription factor |
TF Family: Orphans |
Salt-tolerance protein, putative [Ricinus communis] |
| 6 |
Hb_061256_010 |
0.1913278415 |
- |
- |
PREDICTED: uncharacterized protein LOC105635220 isoform X2 [Jatropha curcas] |
| 7 |
Hb_082655_010 |
0.1920361969 |
- |
- |
rhodanese-like domain-containing family protein [Populus trichocarpa] |
| 8 |
Hb_000123_240 |
0.1920533462 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 9 |
Hb_000235_070 |
0.1940630199 |
- |
- |
PREDICTED: VIN3-like protein 2 [Jatropha curcas] |
| 10 |
Hb_004545_120 |
0.1947951744 |
- |
- |
PREDICTED: uncharacterized protein LOC105647604 [Jatropha curcas] |
| 11 |
Hb_001269_370 |
0.1960294856 |
- |
- |
PREDICTED: U-box domain-containing protein 43 [Jatropha curcas] |
| 12 |
Hb_000334_050 |
0.1973704338 |
- |
- |
PREDICTED: vesicle-associated protein 4-1-like [Jatropha curcas] |
| 13 |
Hb_000270_600 |
0.1976772228 |
- |
- |
PREDICTED: uncharacterized protein LOC105640430 isoform X1 [Jatropha curcas] |
| 14 |
Hb_166876_010 |
0.2007112297 |
- |
- |
xenotropic and polytropic murine leukemia virus receptor ids-4, putative [Ricinus communis] |
| 15 |
Hb_017895_060 |
0.2012192835 |
- |
- |
PREDICTED: rhodanese-like domain-containing protein 14, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000032_510 |
0.2024679634 |
- |
- |
PREDICTED: SUN domain-containing protein 2 [Jatropha curcas] |
| 17 |
Hb_005000_080 |
0.2028783695 |
- |
- |
PREDICTED: thioredoxin-like 2, chloroplastic [Populus euphratica] |
| 18 |
Hb_000011_560 |
0.2058036913 |
- |
- |
PREDICTED: phosphoinositide phosphatase SAC2 isoform X1 [Jatropha curcas] |
| 19 |
Hb_005217_030 |
0.2058834975 |
- |
- |
PREDICTED: probable transcriptional regulator SLK3 [Jatropha curcas] |
| 20 |
Hb_000398_170 |
0.2063905473 |
- |
- |
PREDICTED: tobamovirus multiplication protein 1-like [Jatropha curcas] |