| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
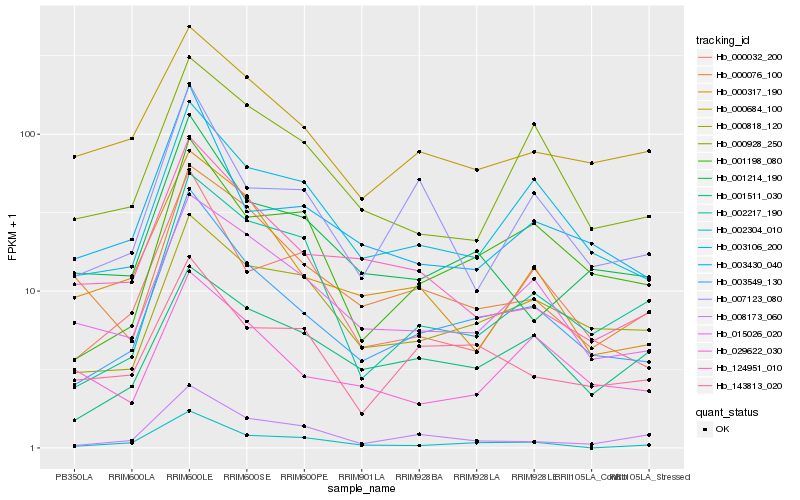
Hb_003549_130 |
0.0 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |
| 2 |
Hb_003106_200 |
0.1267027099 |
- |
- |
PREDICTED: uncharacterized protein LOC105640430 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001198_080 |
0.1380453318 |
- |
- |
PREDICTED: uncharacterized protein LOC105633342 isoform X2 [Jatropha curcas] |
| 4 |
Hb_015026_020 |
0.1410812009 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os07g0563300 [Jatropha curcas] |
| 5 |
Hb_003430_040 |
0.1459136629 |
- |
- |
spermidine synthase 1, putative [Ricinus communis] |
| 6 |
Hb_000032_200 |
0.1498158997 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 7 |
Hb_000818_120 |
0.1510541797 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001214_190 |
0.1528755604 |
- |
- |
hypothetical protein JCGZ_25001 [Jatropha curcas] |
| 9 |
Hb_007123_080 |
0.1579087279 |
- |
- |
big map kinase/bmk, putative [Ricinus communis] |
| 10 |
Hb_002304_010 |
0.157974956 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 11 |
Hb_008173_060 |
0.1579798873 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 2 [Jatropha curcas] |
| 12 |
Hb_143813_020 |
0.1581135423 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0006s10950g [Populus trichocarpa] |
| 13 |
Hb_000684_100 |
0.158589746 |
- |
- |
PREDICTED: cell number regulator 8 [Jatropha curcas] |
| 14 |
Hb_000317_190 |
0.1606976861 |
- |
- |
PREDICTED: diacylglycerol kinase 5-like [Jatropha curcas] |
| 15 |
Hb_000928_250 |
0.1622231343 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 16 |
Hb_000076_100 |
0.1649365951 |
- |
- |
PREDICTED: septum-promoting GTP-binding protein 1 [Jatropha curcas] |
| 17 |
Hb_001511_030 |
0.1649948107 |
- |
- |
PREDICTED: inactive receptor-like serine/threonine-protein kinase At2g40270 [Jatropha curcas] |
| 18 |
Hb_124951_010 |
0.1655217862 |
- |
- |
chitinase-like protein [Hevea brasiliensis] |
| 19 |
Hb_002217_190 |
0.168331206 |
- |
- |
invertase [Hevea brasiliensis] |
| 20 |
Hb_029622_030 |
0.1685197437 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |