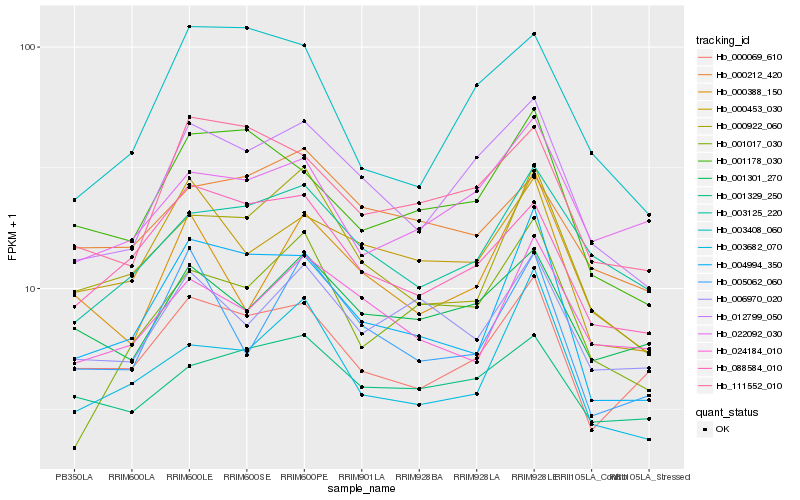
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012799_050 |
0.0 |
- |
- |
PREDICTED: remorin [Jatropha curcas] |
| 2 |
Hb_111552_010 |
0.101257281 |
- |
- |
PREDICTED: ATP sulfurylase 2 [Jatropha curcas] |
| 3 |
Hb_003125_220 |
0.1039931677 |
- |
- |
PREDICTED: CBS domain-containing protein CBSCBSPB1 [Jatropha curcas] |
| 4 |
Hb_000453_030 |
0.1058732906 |
- |
- |
Protein kinase capable of phosphorylating tyrosine family protein [Populus trichocarpa] |
| 5 |
Hb_088584_010 |
0.1096042486 |
- |
- |
hypothetical protein POPTR_0016s08470g [Populus trichocarpa] |
| 6 |
Hb_001301_270 |
0.1102464959 |
- |
- |
PREDICTED: heat shock 70 kDa protein 16 [Jatropha curcas] |
| 7 |
Hb_003408_060 |
0.1134566458 |
transcription factor |
TF Family: Orphans |
Salt-tolerance protein, putative [Ricinus communis] |
| 8 |
Hb_001178_030 |
0.1162486807 |
- |
- |
PREDICTED: 1,4-alpha-glucan-branching enzyme 2-2, chloroplastic/amyloplastic [Jatropha curcas] |
| 9 |
Hb_000922_060 |
0.119028719 |
- |
- |
kinesin light chain, putative [Ricinus communis] |
| 10 |
Hb_000069_610 |
0.1205016949 |
- |
- |
PREDICTED: protein SUPPRESSOR OF PHYA-105 1 [Jatropha curcas] |
| 11 |
Hb_001329_250 |
0.1213484503 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105642692 isoform X1 [Jatropha curcas] |
| 12 |
Hb_004994_350 |
0.1215210694 |
- |
- |
PREDICTED: UPF0187 protein At3g61320, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000388_150 |
0.1219717933 |
- |
- |
hypothetical protein JCGZ_21651 [Jatropha curcas] |
| 14 |
Hb_001017_030 |
0.1219959863 |
- |
- |
copine, putative [Ricinus communis] |
| 15 |
Hb_022092_030 |
0.1237619165 |
- |
- |
PREDICTED: lipoyl synthase, mitochondrial [Jatropha curcas] |
| 16 |
Hb_003682_070 |
0.1265434216 |
- |
- |
PREDICTED: uncharacterized protein LOC105633817 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000212_420 |
0.1267485349 |
- |
- |
PREDICTED: probable protein phosphatase 2C 33 [Jatropha curcas] |
| 18 |
Hb_005062_060 |
0.1288617018 |
- |
- |
PREDICTED: uncharacterized protein At4g06598-like [Jatropha curcas] |
| 19 |
Hb_024184_010 |
0.1290491127 |
- |
- |
PREDICTED: clustered mitochondria protein-like isoform X1 [Citrus sinensis] |
| 20 |
Hb_006970_020 |
0.1297749292 |
- |
- |
PREDICTED: DNA topoisomerase 6 subunit B [Jatropha curcas] |