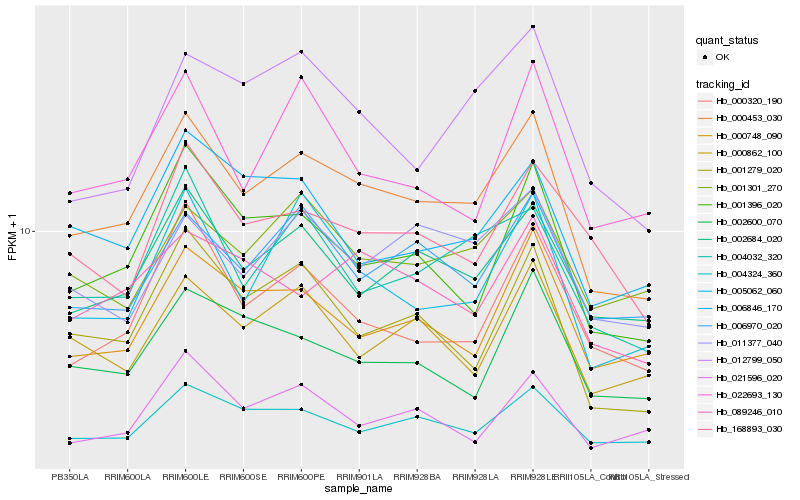
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000453_030 |
0.0 |
- |
- |
Protein kinase capable of phosphorylating tyrosine family protein [Populus trichocarpa] |
| 2 |
Hb_002684_020 |
0.0820978301 |
- |
- |
ATP-dependent clp protease ATP-binding subunit clpx, putative [Ricinus communis] |
| 3 |
Hb_001279_020 |
0.0910735067 |
- |
- |
PREDICTED: uncharacterized protein LOC105633240 [Jatropha curcas] |
| 4 |
Hb_001396_020 |
0.0924853791 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 5 |
Hb_006970_020 |
0.0942134363 |
- |
- |
PREDICTED: DNA topoisomerase 6 subunit B [Jatropha curcas] |
| 6 |
Hb_005062_060 |
0.0949542244 |
- |
- |
PREDICTED: uncharacterized protein At4g06598-like [Jatropha curcas] |
| 7 |
Hb_004032_320 |
0.0953744229 |
- |
- |
PREDICTED: uncharacterized protein LOC105635153 [Jatropha curcas] |
| 8 |
Hb_000748_090 |
0.0966794017 |
- |
- |
glucose inhibited division protein A, putative [Ricinus communis] |
| 9 |
Hb_004324_360 |
0.098927446 |
- |
- |
PREDICTED: uncharacterized protein LOC105650600 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000320_190 |
0.1011334286 |
- |
- |
PREDICTED: aminomethyltransferase, mitochondrial [Jatropha curcas] |
| 11 |
Hb_001301_270 |
0.102247137 |
- |
- |
PREDICTED: heat shock 70 kDa protein 16 [Jatropha curcas] |
| 12 |
Hb_000862_100 |
0.1024285846 |
- |
- |
PREDICTED: plastid division protein CDP1, chloroplastic [Jatropha curcas] |
| 13 |
Hb_089246_010 |
0.1045869703 |
- |
- |
hypothetical protein JCGZ_24395 [Jatropha curcas] |
| 14 |
Hb_011377_040 |
0.1058679326 |
- |
- |
PREDICTED: uncharacterized protein LOC105647286 [Jatropha curcas] |
| 15 |
Hb_012799_050 |
0.1058732906 |
- |
- |
PREDICTED: remorin [Jatropha curcas] |
| 16 |
Hb_002600_070 |
0.1071214237 |
transcription factor |
TF Family: TAZ |
PREDICTED: histone acetyltransferase HAC12 [Jatropha curcas] |
| 17 |
Hb_021596_020 |
0.1072194748 |
- |
- |
hypothetical protein JCGZ_02034 [Jatropha curcas] |
| 18 |
Hb_168893_030 |
0.1077932384 |
- |
- |
PREDICTED: U-box domain-containing protein 13-like [Jatropha curcas] |
| 19 |
Hb_022693_130 |
0.1081969882 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 20 |
Hb_006846_170 |
0.1088233663 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX10L [Jatropha curcas] |