| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
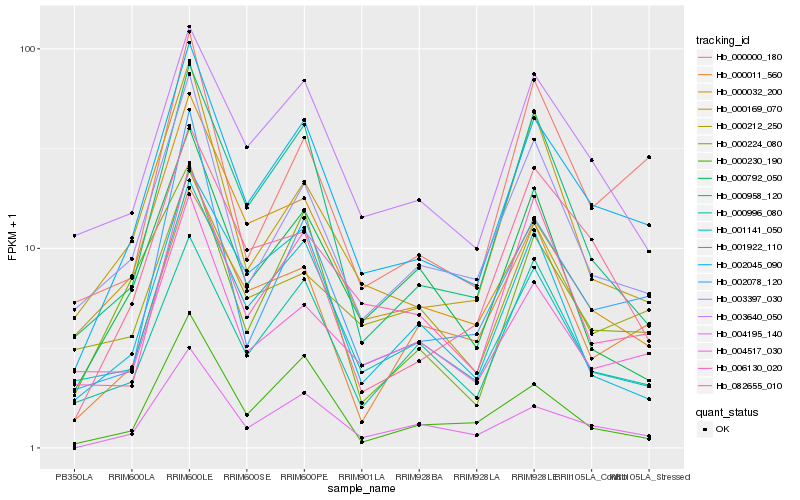
Hb_002045_090 |
0.0 |
- |
- |
PREDICTED: tryptophan synthase alpha chain-like [Jatropha curcas] |
| 2 |
Hb_004195_140 |
0.1110391562 |
- |
- |
Sec14 cytosolic factor, putative [Ricinus communis] |
| 3 |
Hb_003640_050 |
0.1205488834 |
- |
- |
PREDICTED: protein LUTEIN DEFICIENT 5, chloroplastic isoform X2 [Jatropha curcas] |
| 4 |
Hb_000958_120 |
0.1262276642 |
- |
- |
PREDICTED: uncharacterized protein LOC105646385 [Jatropha curcas] |
| 5 |
Hb_003397_030 |
0.1290105711 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit beta-3, chloroplastic [Jatropha curcas] |
| 6 |
Hb_001141_050 |
0.129600025 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 2 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000224_080 |
0.1299413381 |
- |
- |
hypothetical protein RCOM_0510880 [Ricinus communis] |
| 8 |
Hb_002078_120 |
0.1358046311 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g67480 [Jatropha curcas] |
| 9 |
Hb_000996_080 |
0.1410116841 |
- |
- |
PREDICTED: intracellular protein transport protein USO1 [Jatropha curcas] |
| 10 |
Hb_082655_010 |
0.1437050436 |
- |
- |
rhodanese-like domain-containing family protein [Populus trichocarpa] |
| 11 |
Hb_000000_180 |
0.145371582 |
- |
- |
PREDICTED: 30S ribosomal protein S9, chloroplastic-like [Populus euphratica] |
| 12 |
Hb_004517_030 |
0.1470177116 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000169_070 |
0.14796314 |
- |
- |
PREDICTED: protein TRANSPORT INHIBITOR RESPONSE 1 [Jatropha curcas] |
| 14 |
Hb_006130_020 |
0.149803887 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 15 |
Hb_000230_190 |
0.1501006173 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000792_050 |
0.1503626358 |
- |
- |
PREDICTED: acetyl-coenzyme A carboxylase carboxyl transferase subunit alpha, chloroplastic isoform X1 [Jatropha curcas] |
| 17 |
Hb_000011_560 |
0.1509242735 |
- |
- |
PREDICTED: phosphoinositide phosphatase SAC2 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001922_110 |
0.1511681921 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000212_250 |
0.1516810557 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000032_200 |
0.1523349351 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |