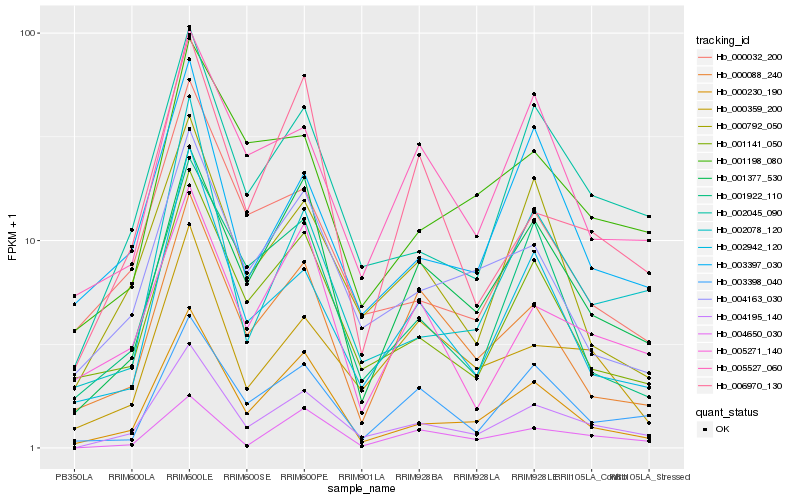
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004195_140 |
0.0 |
- |
- |
Sec14 cytosolic factor, putative [Ricinus communis] |
| 2 |
Hb_002045_090 |
0.1110391562 |
- |
- |
PREDICTED: tryptophan synthase alpha chain-like [Jatropha curcas] |
| 3 |
Hb_000230_190 |
0.1304018858 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK2 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001377_530 |
0.135843202 |
- |
- |
NC domain-containing family protein [Populus trichocarpa] |
| 5 |
Hb_001141_050 |
0.1429098602 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 2 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000792_050 |
0.1520305832 |
- |
- |
PREDICTED: acetyl-coenzyme A carboxylase carboxyl transferase subunit alpha, chloroplastic isoform X1 [Jatropha curcas] |
| 7 |
Hb_002942_120 |
0.1524375872 |
transcription factor |
TF Family: bHLH |
Transcription factor ICE1, putative [Ricinus communis] |
| 8 |
Hb_000359_200 |
0.1564746309 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 9 |
Hb_004163_030 |
0.1570050145 |
- |
- |
PREDICTED: protein PHLOEM PROTEIN 2-LIKE A10 [Jatropha curcas] |
| 10 |
Hb_003398_040 |
0.1575045093 |
- |
- |
PREDICTED: actin-related protein 5 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002078_120 |
0.1587615883 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g67480 [Jatropha curcas] |
| 12 |
Hb_005527_060 |
0.1636311285 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 13 |
Hb_004650_030 |
0.1640662927 |
- |
- |
PREDICTED: uncharacterized protein LOC105638888 isoform X1 [Jatropha curcas] |
| 14 |
Hb_003397_030 |
0.1655596152 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit beta-3, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000032_200 |
0.1657199878 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 16 |
Hb_005271_140 |
0.1664576035 |
transcription factor |
TF Family: LIM |
Pollen-specific protein SF3 [Morus notabilis] |
| 17 |
Hb_001198_080 |
0.1671682298 |
- |
- |
PREDICTED: uncharacterized protein LOC105633342 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000088_240 |
0.1677617159 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 19 |
Hb_001922_110 |
0.1680937268 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_006970_130 |
0.1689596058 |
- |
- |
PREDICTED: histone H3.3-like [Colobus angolensis palliatus] |