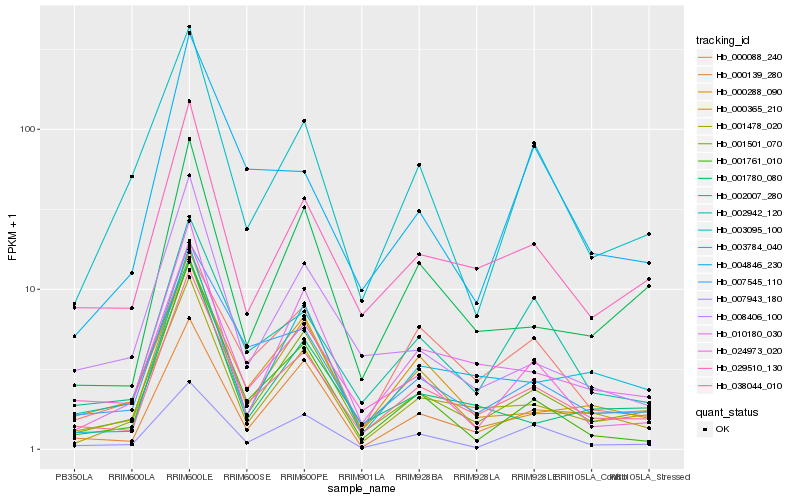
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_038044_010 |
0.0 |
- |
- |
12-oxophytodienoate reductase 2 [Theobroma cacao] |
| 2 |
Hb_001501_070 |
0.0863042308 |
- |
- |
PREDICTED: condensin complex subunit 2 [Jatropha curcas] |
| 3 |
Hb_029510_130 |
0.0898380153 |
- |
- |
PREDICTED: malonate--CoA ligase [Jatropha curcas] |
| 4 |
Hb_007545_110 |
0.1055797382 |
- |
- |
catalytic, putative [Ricinus communis] |
| 5 |
Hb_024973_020 |
0.106156138 |
- |
- |
Protein regulator of cytokinesis, putative [Ricinus communis] |
| 6 |
Hb_001478_020 |
0.1067219866 |
desease resistance |
Gene Name: AAA |
thyroid hormone receptor interactor, putative [Ricinus communis] |
| 7 |
Hb_002942_120 |
0.121347851 |
transcription factor |
TF Family: bHLH |
Transcription factor ICE1, putative [Ricinus communis] |
| 8 |
Hb_010180_030 |
0.1257657608 |
- |
- |
PREDICTED: WEB family protein At2g38370-like [Jatropha curcas] |
| 9 |
Hb_000365_210 |
0.131084431 |
- |
- |
PREDICTED: cell division control protein 2 homolog C [Jatropha curcas] |
| 10 |
Hb_001780_080 |
0.1359197278 |
- |
- |
PREDICTED: tubulin beta-2 chain [Jatropha curcas] |
| 11 |
Hb_007943_180 |
0.1386369062 |
- |
- |
PREDICTED: uncharacterized protein LOC105637734 [Jatropha curcas] |
| 12 |
Hb_001761_010 |
0.1386794113 |
- |
- |
PREDICTED: uncharacterized protein LOC105639282 [Jatropha curcas] |
| 13 |
Hb_000288_090 |
0.1421727218 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 14 |
Hb_003784_040 |
0.1458100431 |
- |
- |
Afadin- and alpha-actinin-binding protein, putative [Ricinus communis] |
| 15 |
Hb_003095_100 |
0.1479880154 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_008406_100 |
0.1504612903 |
- |
- |
PREDICTED: protein TPX2 isoform X2 [Jatropha curcas] |
| 17 |
Hb_004846_230 |
0.1522247898 |
- |
- |
Chaperone protein dnaJ 8, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_000088_240 |
0.1527649772 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 19 |
Hb_002007_280 |
0.1530263609 |
- |
- |
PREDICTED: kinesin-like protein KIFC3 [Jatropha curcas] |
| 20 |
Hb_000139_280 |
0.153896088 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase NPK1 isoform X1 [Jatropha curcas] |