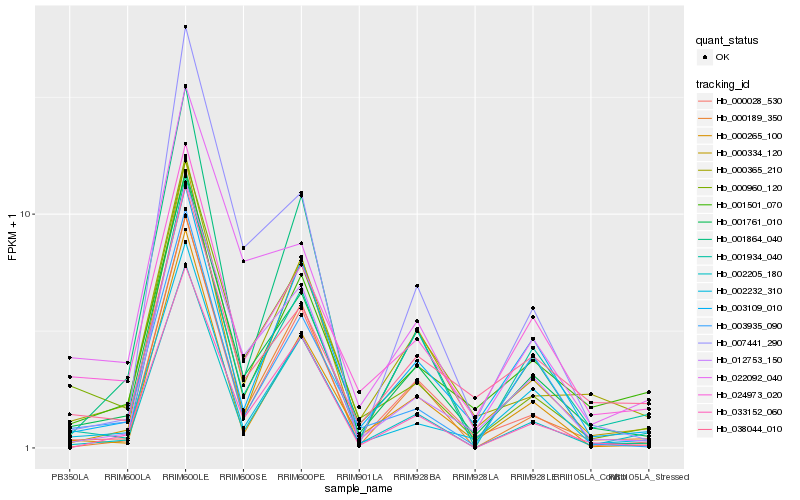
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001761_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105639282 [Jatropha curcas] |
| 2 |
Hb_012753_150 |
0.1007809102 |
transcription factor |
TF Family: GRAS |
DELLA protein RGL1, putative [Ricinus communis] |
| 3 |
Hb_000028_530 |
0.1079426973 |
- |
- |
Protein regulator of cytokinesis, putative [Ricinus communis] |
| 4 |
Hb_000365_210 |
0.1079920774 |
- |
- |
PREDICTED: cell division control protein 2 homolog C [Jatropha curcas] |
| 5 |
Hb_000960_120 |
0.1108431818 |
- |
- |
PREDICTED: cell division cycle 20.1, cofactor of APC complex-like [Jatropha curcas] |
| 6 |
Hb_002232_310 |
0.1111753854 |
- |
- |
PREDICTED: uncharacterized protein LOC105635987 [Jatropha curcas] |
| 7 |
Hb_024973_020 |
0.1189765452 |
- |
- |
Protein regulator of cytokinesis, putative [Ricinus communis] |
| 8 |
Hb_000265_100 |
0.1201165304 |
- |
- |
PREDICTED: uncharacterized protein LOC105643473 [Jatropha curcas] |
| 9 |
Hb_022092_040 |
0.1224308978 |
- |
- |
PREDICTED: serine/arginine repetitive matrix protein 1 [Jatropha curcas] |
| 10 |
Hb_001501_070 |
0.1245197073 |
- |
- |
PREDICTED: condensin complex subunit 2 [Jatropha curcas] |
| 11 |
Hb_001934_040 |
0.1268308059 |
- |
- |
PREDICTED: vacuolar iron transporter 1-like [Jatropha curcas] |
| 12 |
Hb_007441_290 |
0.1295928562 |
- |
- |
PREDICTED: uncharacterized protein LOC105643262 [Jatropha curcas] |
| 13 |
Hb_033152_060 |
0.1298948393 |
- |
- |
PREDICTED: phragmoplast orienting kinesin 2 [Jatropha curcas] |
| 14 |
Hb_000334_120 |
0.130714763 |
- |
- |
PREDICTED: DNA topoisomerase 2 [Jatropha curcas] |
| 15 |
Hb_003935_090 |
0.1318271831 |
- |
- |
PREDICTED: uncharacterized protein At4g38062 [Jatropha curcas] |
| 16 |
Hb_000189_350 |
0.1343921454 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 3 [Jatropha curcas] |
| 17 |
Hb_003109_010 |
0.1348604603 |
- |
- |
PREDICTED: kinesin-like protein KIN12B [Jatropha curcas] |
| 18 |
Hb_001864_040 |
0.1353715646 |
- |
- |
PREDICTED: kinesin-1 [Jatropha curcas] |
| 19 |
Hb_002205_180 |
0.1374742538 |
- |
- |
PREDICTED: uncharacterized protein LOC105649178 [Jatropha curcas] |
| 20 |
Hb_038044_010 |
0.1386794113 |
- |
- |
12-oxophytodienoate reductase 2 [Theobroma cacao] |