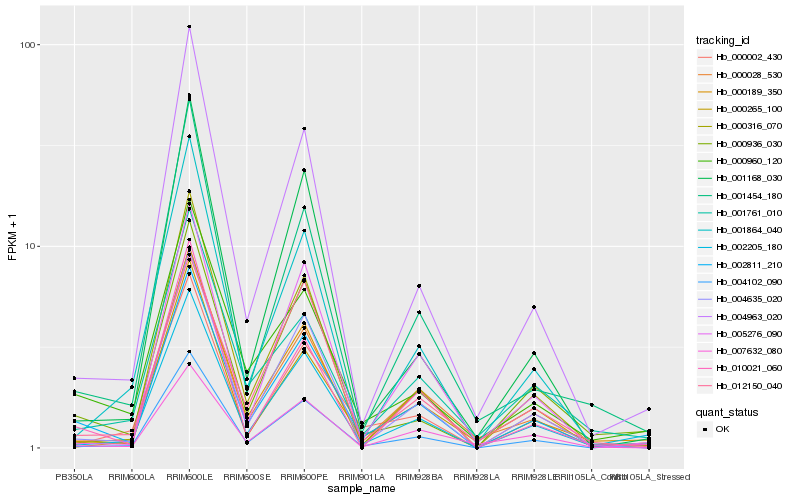
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002811_210 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek1 isoform X1 [Jatropha curcas] |
| 2 |
Hb_004635_020 |
0.0967922003 |
- |
- |
PREDICTED: uncharacterized protein At3g28850-like [Jatropha curcas] |
| 3 |
Hb_000028_530 |
0.1243584573 |
- |
- |
Protein regulator of cytokinesis, putative [Ricinus communis] |
| 4 |
Hb_000960_120 |
0.1253699099 |
- |
- |
PREDICTED: cell division cycle 20.1, cofactor of APC complex-like [Jatropha curcas] |
| 5 |
Hb_000265_100 |
0.1310408683 |
- |
- |
PREDICTED: uncharacterized protein LOC105643473 [Jatropha curcas] |
| 6 |
Hb_010021_060 |
0.1318422866 |
- |
- |
PREDICTED: kinesin-related protein 11 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000936_030 |
0.1331265769 |
- |
- |
hypothetical protein RCOM_0615680 [Ricinus communis] |
| 8 |
Hb_000316_070 |
0.1334376897 |
- |
- |
PREDICTED: actin-1-like isoform X2 [Cucumis melo] |
| 9 |
Hb_004102_090 |
0.1346136442 |
transcription factor |
TF Family: CPP |
hypothetical protein JCGZ_06186 [Jatropha curcas] |
| 10 |
Hb_007632_080 |
0.1350024532 |
- |
- |
PREDICTED: intracellular protein transport protein USO1 [Jatropha curcas] |
| 11 |
Hb_000002_430 |
0.1365704177 |
- |
- |
PREDICTED: FHA domain-containing protein At4g14490 [Populus euphratica] |
| 12 |
Hb_004963_020 |
0.1374495951 |
- |
- |
PREDICTED: patellin-4 [Populus euphratica] |
| 13 |
Hb_001454_180 |
0.1431229637 |
- |
- |
PREDICTED: uncharacterized protein LOC105643432 [Jatropha curcas] |
| 14 |
Hb_001864_040 |
0.1434792355 |
- |
- |
PREDICTED: kinesin-1 [Jatropha curcas] |
| 15 |
Hb_002205_180 |
0.1438018485 |
- |
- |
PREDICTED: uncharacterized protein LOC105649178 [Jatropha curcas] |
| 16 |
Hb_000189_350 |
0.145993865 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 3 [Jatropha curcas] |
| 17 |
Hb_005276_090 |
0.1465555136 |
- |
- |
PREDICTED: uncharacterized protein LOC105649997 isoform X2 [Jatropha curcas] |
| 18 |
Hb_001761_010 |
0.1480924251 |
- |
- |
PREDICTED: uncharacterized protein LOC105639282 [Jatropha curcas] |
| 19 |
Hb_001168_030 |
0.1481264307 |
- |
- |
PREDICTED: G2/mitotic-specific cyclin S13-7 [Jatropha curcas] |
| 20 |
Hb_012150_040 |
0.1483985977 |
- |
- |
conserved hypothetical protein [Ricinus communis] |