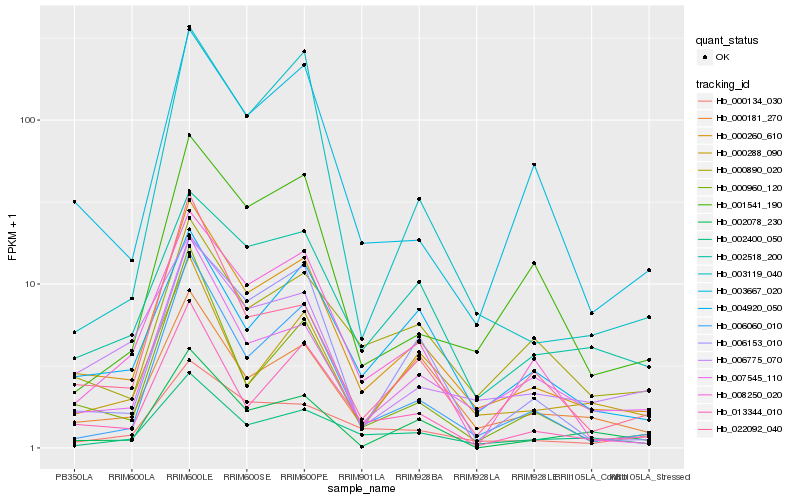
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000260_610 |
0.0 |
- |
- |
PREDICTED: cell division cycle 20.1, cofactor of APC complex-like [Jatropha curcas] |
| 2 |
Hb_013344_010 |
0.1215195101 |
- |
- |
PREDICTED: microtubule-associated protein TORTIFOLIA1 [Jatropha curcas] |
| 3 |
Hb_006060_010 |
0.1236973623 |
- |
- |
PREDICTED: cyclin-A2-2-like [Jatropha curcas] |
| 4 |
Hb_007545_110 |
0.1273478104 |
- |
- |
catalytic, putative [Ricinus communis] |
| 5 |
Hb_002400_050 |
0.1394739694 |
- |
- |
PREDICTED: uncharacterized protein LOC105637632 isoform X1 [Jatropha curcas] |
| 6 |
Hb_003667_020 |
0.1401069967 |
- |
- |
conserved hypothetical protein 16 [Hevea brasiliensis] |
| 7 |
Hb_004920_050 |
0.140685274 |
- |
- |
DNA replication licensing factor MCM7, putative [Ricinus communis] |
| 8 |
Hb_002518_200 |
0.1424328743 |
- |
- |
Proliferating cell nuclear antigen [Theobroma cacao] |
| 9 |
Hb_008250_020 |
0.1430758041 |
- |
- |
PREDICTED: U-box domain-containing protein 15-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_006153_010 |
0.1432975215 |
- |
- |
PREDICTED: monocopper oxidase-like protein SKU5 [Populus euphratica] |
| 11 |
Hb_000960_120 |
0.1447475558 |
- |
- |
PREDICTED: cell division cycle 20.1, cofactor of APC complex-like [Jatropha curcas] |
| 12 |
Hb_000134_030 |
0.1468232147 |
- |
- |
PREDICTED: sister chromatid cohesion 1 protein 3-like [Jatropha curcas] |
| 13 |
Hb_002078_230 |
0.1477500378 |
- |
- |
PREDICTED: HORMA domain-containing protein 1 [Jatropha curcas] |
| 14 |
Hb_022092_040 |
0.1479787759 |
- |
- |
PREDICTED: serine/arginine repetitive matrix protein 1 [Jatropha curcas] |
| 15 |
Hb_000890_020 |
0.1504145896 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |
| 16 |
Hb_000181_270 |
0.1517647167 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 17 |
Hb_000288_090 |
0.1530135162 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_006775_070 |
0.1541320898 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 19 |
Hb_001541_190 |
0.1603426706 |
- |
- |
PREDICTED: uncharacterized protein LOC105644700 [Jatropha curcas] |
| 20 |
Hb_003119_040 |
0.1705473938 |
- |
- |
PREDICTED: classical arabinogalactan protein 1-like [Populus euphratica] |