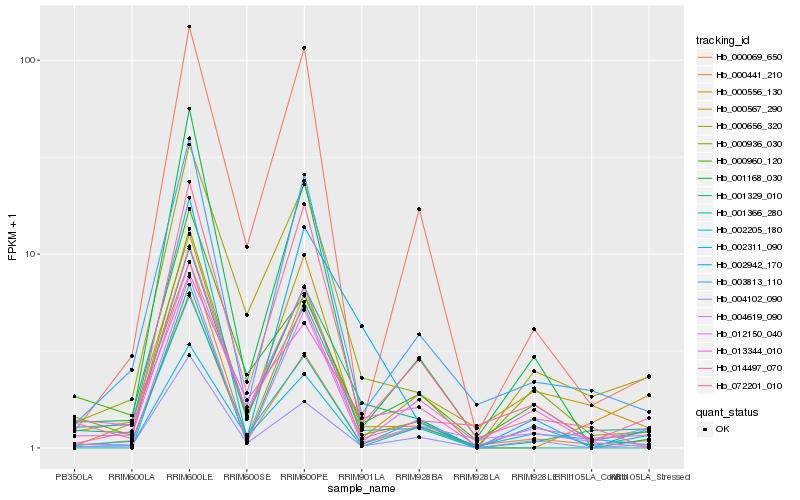
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001329_010 |
0.0 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 2 |
Hb_000656_320 |
0.1205554718 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit gamma 3-like [Jatropha curcas] |
| 3 |
Hb_013344_010 |
0.1424002882 |
- |
- |
PREDICTED: microtubule-associated protein TORTIFOLIA1 [Jatropha curcas] |
| 4 |
Hb_000556_130 |
0.143556189 |
- |
- |
PREDICTED: serine/threonine-protein kinase Aurora-3 [Jatropha curcas] |
| 5 |
Hb_003813_110 |
0.1569232306 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002942_170 |
0.1598497655 |
- |
- |
acyl carrier protein, putative [Ricinus communis] |
| 7 |
Hb_072201_010 |
0.1599103256 |
- |
- |
PREDICTED: cell division control protein 2 homolog C [Jatropha curcas] |
| 8 |
Hb_000441_210 |
0.1619148482 |
- |
- |
PREDICTED: uncharacterized protein LOC105636842 [Jatropha curcas] |
| 9 |
Hb_000936_030 |
0.1670645974 |
- |
- |
hypothetical protein RCOM_0615680 [Ricinus communis] |
| 10 |
Hb_004619_090 |
0.1679205533 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002205_180 |
0.1870448945 |
- |
- |
PREDICTED: uncharacterized protein LOC105649178 [Jatropha curcas] |
| 12 |
Hb_004102_090 |
0.1871572198 |
transcription factor |
TF Family: CPP |
hypothetical protein JCGZ_06186 [Jatropha curcas] |
| 13 |
Hb_012150_040 |
0.1890786447 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000069_650 |
0.1895559982 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000960_120 |
0.1916199875 |
- |
- |
PREDICTED: cell division cycle 20.1, cofactor of APC complex-like [Jatropha curcas] |
| 16 |
Hb_002311_090 |
0.1950017031 |
- |
- |
PREDICTED: uncharacterized protein LOC105643503 [Jatropha curcas] |
| 17 |
Hb_014497_070 |
0.1951502052 |
- |
- |
PREDICTED: cyclin-dependent kinase B2-2 [Jatropha curcas] |
| 18 |
Hb_001366_280 |
0.1951561536 |
- |
- |
PREDICTED: uncharacterized protein LOC105642732 [Jatropha curcas] |
| 19 |
Hb_000567_290 |
0.1963571664 |
- |
- |
hypothetical protein RCOM_0303940 [Ricinus communis] |
| 20 |
Hb_001168_030 |
0.1968435602 |
- |
- |
PREDICTED: G2/mitotic-specific cyclin S13-7 [Jatropha curcas] |