| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
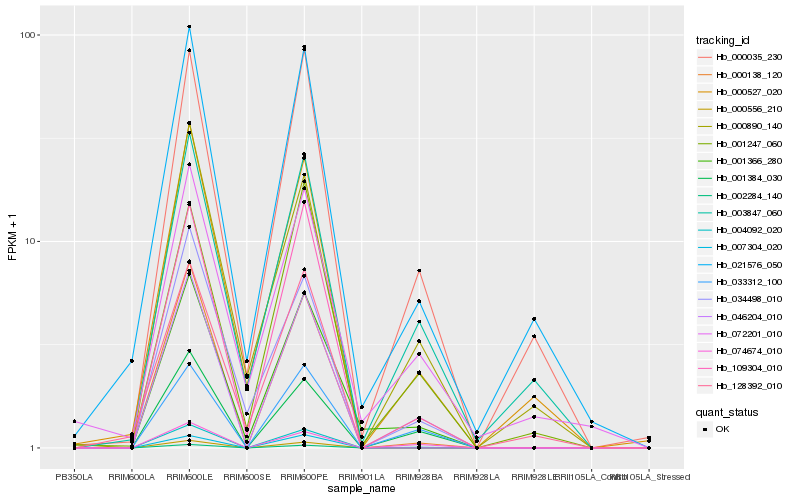
Hb_001366_280 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642732 [Jatropha curcas] |
| 2 |
Hb_046204_010 |
0.1208896519 |
- |
- |
PREDICTED: peroxidase 5-like [Jatropha curcas] |
| 3 |
Hb_007304_020 |
0.1342871157 |
- |
- |
PREDICTED: uncharacterized protein LOC100256879 [Vitis vinifera] |
| 4 |
Hb_021576_050 |
0.1381303196 |
- |
- |
PREDICTED: early nodulin-like protein 1 [Jatropha curcas] |
| 5 |
Hb_000035_230 |
0.1401454345 |
- |
- |
PREDICTED: early nodulin-like protein 1 [Jatropha curcas] |
| 6 |
Hb_000138_120 |
0.1403423841 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_034498_010 |
0.1445693121 |
- |
- |
PREDICTED: uncharacterized protein LOC105636842 [Jatropha curcas] |
| 8 |
Hb_000527_020 |
0.1452368564 |
- |
- |
PREDICTED: uncharacterized protein LOC105649490 [Jatropha curcas] |
| 9 |
Hb_072201_010 |
0.1485990556 |
- |
- |
PREDICTED: cell division control protein 2 homolog C [Jatropha curcas] |
| 10 |
Hb_128392_010 |
0.1508161077 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein At3g19184 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001384_030 |
0.1516649795 |
- |
- |
PREDICTED: uncharacterized protein LOC105049010 [Elaeis guineensis] |
| 12 |
Hb_001247_060 |
0.1519012771 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 13 |
Hb_003847_060 |
0.1526457517 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 14 |
Hb_033312_100 |
0.1564658873 |
- |
- |
- |
| 15 |
Hb_000890_140 |
0.1566381804 |
- |
- |
PREDICTED: uncharacterized protein LOC105646185 [Jatropha curcas] |
| 16 |
Hb_074674_010 |
0.158108907 |
- |
- |
Cell division cycle protein cdt2, putative [Ricinus communis] |
| 17 |
Hb_004092_020 |
0.1602210261 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 32 [Jatropha curcas] |
| 18 |
Hb_002284_140 |
0.1602227188 |
- |
- |
hypothetical protein POPTR_0002s00800g [Populus trichocarpa] |
| 19 |
Hb_000556_210 |
0.1603296111 |
- |
- |
hypothetical protein POPTR_0014s06640g [Populus trichocarpa] |
| 20 |
Hb_109304_010 |
0.1604303997 |
- |
- |
hypothetical protein JCGZ_19971 [Jatropha curcas] |