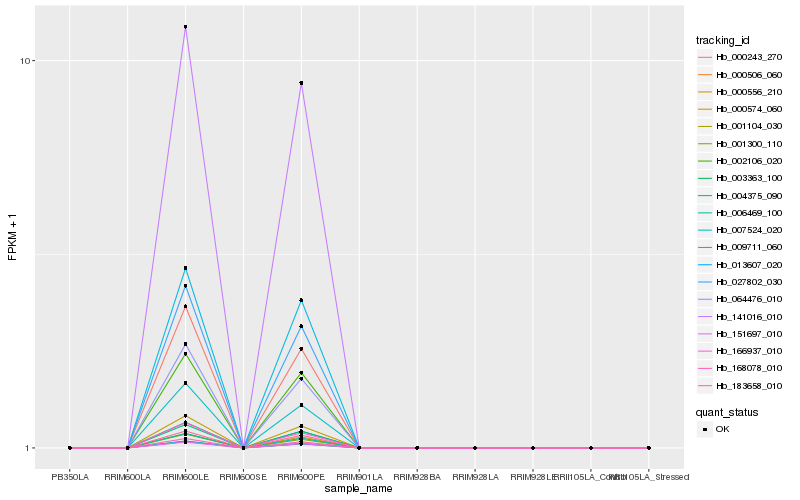
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006469_100 |
0.0 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Malus domestica] |
| 2 |
Hb_000574_060 |
0.0015452763 |
- |
- |
hypothetical protein F775_19731 [Aegilops tauschii] |
| 3 |
Hb_027802_030 |
0.0026083391 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0017s11800g [Populus trichocarpa] |
| 4 |
Hb_168078_010 |
0.005300993 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g62930, chloroplastic-like, partial [Jatropha curcas] |
| 5 |
Hb_003363_100 |
0.0083291928 |
- |
- |
- |
| 6 |
Hb_141016_010 |
0.008486005 |
- |
- |
hypothetical protein JCGZ_15085 [Jatropha curcas] |
| 7 |
Hb_000506_060 |
0.0106901016 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: subtilisin-like protease [Malus domestica] |
| 8 |
Hb_004375_090 |
0.0119106048 |
- |
- |
hypothetical protein JCGZ_13971 [Jatropha curcas] |
| 9 |
Hb_007524_020 |
0.0142931689 |
- |
- |
PREDICTED: probable indole-3-pyruvate monooxygenase YUCCA4 [Jatropha curcas] |
| 10 |
Hb_001104_030 |
0.0151735259 |
- |
- |
PREDICTED: uncharacterized protein LOC105125581 [Populus euphratica] |
| 11 |
Hb_001300_110 |
0.0165120118 |
- |
- |
PREDICTED: bidirectional sugar transporter N3-like [Jatropha curcas] |
| 12 |
Hb_000243_270 |
0.0175427883 |
- |
- |
Pectinesterase-2 precursor, putative [Ricinus communis] |
| 13 |
Hb_064476_010 |
0.0215648119 |
- |
- |
PREDICTED: beta-Amyrin Synthase 1-like [Sesamum indicum] |
| 14 |
Hb_009711_060 |
0.0221165424 |
- |
- |
- |
| 15 |
Hb_151697_010 |
0.0223780141 |
- |
- |
clathrin assembly protein, putative [Ricinus communis] |
| 16 |
Hb_183658_010 |
0.0234822051 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 17 |
Hb_013607_020 |
0.0255627466 |
transcription factor |
TF Family: bZIP |
DNA binding protein, putative [Ricinus communis] |
| 18 |
Hb_002106_020 |
0.0264358672 |
- |
- |
PREDICTED: gibberellic acid methyltransferase 2 [Vitis vinifera] |
| 19 |
Hb_166937_010 |
0.027163653 |
- |
- |
PREDICTED: formyltetrahydrofolate deformylase 1, mitochondrial-like [Jatropha curcas] |
| 20 |
Hb_000556_210 |
0.0290214078 |
- |
- |
hypothetical protein POPTR_0014s06640g [Populus trichocarpa] |