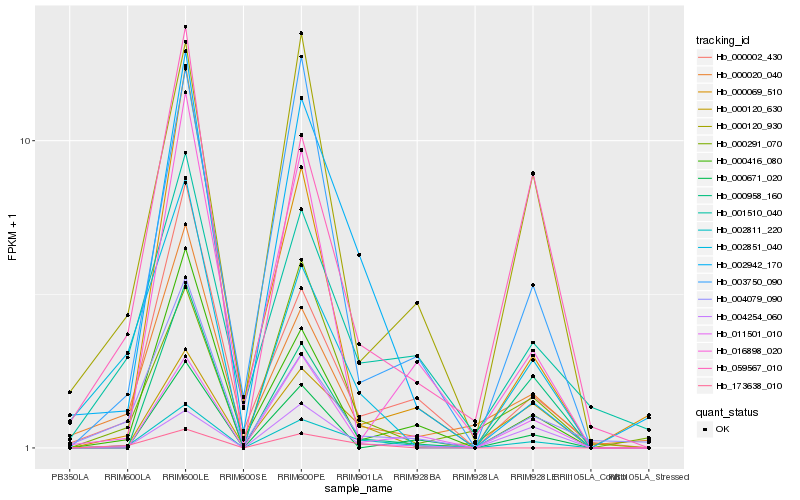
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002811_220 |
0.0 |
transcription factor |
TF Family: Trihelix |
transcription factor, putative [Ricinus communis] |
| 2 |
Hb_002942_170 |
0.1693644067 |
- |
- |
acyl carrier protein, putative [Ricinus communis] |
| 3 |
Hb_000002_430 |
0.1808895998 |
- |
- |
PREDICTED: FHA domain-containing protein At4g14490 [Populus euphratica] |
| 4 |
Hb_000416_080 |
0.1881320277 |
- |
- |
Carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase, putative [Ricinus communis] |
| 5 |
Hb_003750_090 |
0.1910031185 |
- |
- |
BnaC02g34020D [Brassica napus] |
| 6 |
Hb_000120_630 |
0.1950912842 |
- |
- |
PREDICTED: glutamate receptor 3.6-like [Jatropha curcas] |
| 7 |
Hb_059567_010 |
0.1989396584 |
- |
- |
PREDICTED: transcription factor PAR2-like [Jatropha curcas] |
| 8 |
Hb_011501_010 |
0.2002123734 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG5-like [Jatropha curcas] |
| 9 |
Hb_004079_090 |
0.2033063822 |
- |
- |
hypothetical protein RCOM_1053120 [Ricinus communis] |
| 10 |
Hb_001510_040 |
0.207663443 |
- |
- |
PREDICTED: 125 kDa kinesin-related protein [Jatropha curcas] |
| 11 |
Hb_002851_040 |
0.2108390702 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 5 [Jatropha curcas] |
| 12 |
Hb_000120_930 |
0.2132578955 |
- |
- |
PREDICTED: G2/mitotic-specific cyclin-2-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_004254_060 |
0.2161559999 |
- |
- |
PREDICTED: uncharacterized protein LOC105650679 [Jatropha curcas] |
| 14 |
Hb_000958_160 |
0.2195239506 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG1-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_000020_040 |
0.2206555801 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 16 |
Hb_000291_070 |
0.2221749297 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_016898_020 |
0.228870352 |
transcription factor |
TF Family: SET |
PREDICTED: uncharacterized protein LOC105120899 [Populus euphratica] |
| 18 |
Hb_173638_010 |
0.2288793041 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 19 |
Hb_000069_510 |
0.2294164692 |
- |
- |
PREDICTED: histone H3-like centromeric protein HTR12 [Jatropha curcas] |
| 20 |
Hb_000671_020 |
0.2302846092 |
- |
- |
PREDICTED: RNA-binding protein 38 [Jatropha curcas] |