| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
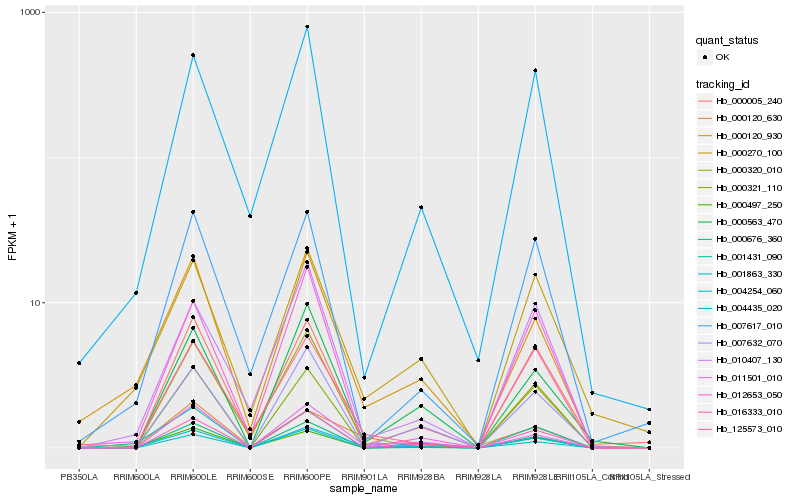
Hb_004254_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105650679 [Jatropha curcas] |
| 2 |
Hb_000120_930 |
0.1426501531 |
- |
- |
PREDICTED: G2/mitotic-specific cyclin-2-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_011501_010 |
0.1482064245 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG5-like [Jatropha curcas] |
| 4 |
Hb_016333_010 |
0.1569084695 |
- |
- |
PREDICTED: probable terpene synthase 13 [Jatropha curcas] |
| 5 |
Hb_000497_250 |
0.1587437679 |
- |
- |
PREDICTED: potassium transporter 5 [Jatropha curcas] |
| 6 |
Hb_001431_090 |
0.1590625305 |
transcription factor |
TF Family: DBP |
PREDICTED: probable protein phosphatase 2C 47, partial [Camelina sativa] |
| 7 |
Hb_000676_360 |
0.1598031297 |
- |
- |
hypothetical protein POPTR_0007s13690g [Populus trichocarpa] |
| 8 |
Hb_000320_010 |
0.1606894343 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_007632_070 |
0.1608636107 |
- |
- |
kinase, putative [Ricinus communis] |
| 10 |
Hb_001863_330 |
0.163764237 |
- |
- |
RNA-binding region-containing protein, putative [Ricinus communis] |
| 11 |
Hb_000270_100 |
0.1652383705 |
- |
- |
ATPase 4, plasma membrane-type -like protein [Gossypium arboreum] |
| 12 |
Hb_125573_010 |
0.165510876 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 13 |
Hb_000005_240 |
0.1694143301 |
- |
- |
Shugoshin-1, putative [Ricinus communis] |
| 14 |
Hb_000563_470 |
0.16980055 |
- |
- |
PREDICTED: uncharacterized protein LOC105643461 [Jatropha curcas] |
| 15 |
Hb_000321_110 |
0.1717233418 |
- |
- |
Uncharacterized protein TCM_041937 [Theobroma cacao] |
| 16 |
Hb_007617_010 |
0.1734937302 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH63 isoform X1 [Jatropha curcas] |
| 17 |
Hb_010407_130 |
0.1735537637 |
- |
- |
hypothetical protein POPTR_0008s16680g [Populus trichocarpa] |
| 18 |
Hb_000120_630 |
0.1756936405 |
- |
- |
PREDICTED: glutamate receptor 3.6-like [Jatropha curcas] |
| 19 |
Hb_004435_020 |
0.1759533037 |
- |
- |
PREDICTED: tubulin alpha-2 chain [Jatropha curcas] |
| 20 |
Hb_012653_050 |
0.1763234015 |
- |
- |
hypothetical protein RCOM_0460020 [Ricinus communis] |