| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
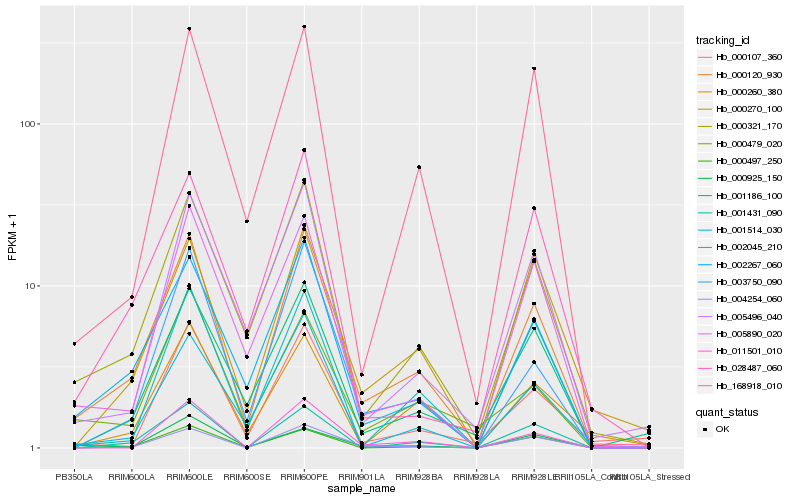
Hb_000120_930 |
0.0 |
- |
- |
PREDICTED: G2/mitotic-specific cyclin-2-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_002267_060 |
0.1113560232 |
transcription factor |
TF Family: ERF |
PREDICTED: dehydration-responsive element-binding protein 3 [Jatropha curcas] |
| 3 |
Hb_000321_170 |
0.111857013 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 11-like [Jatropha curcas] |
| 4 |
Hb_004254_060 |
0.1426501531 |
- |
- |
PREDICTED: uncharacterized protein LOC105650679 [Jatropha curcas] |
| 5 |
Hb_003750_090 |
0.1436518775 |
- |
- |
BnaC02g34020D [Brassica napus] |
| 6 |
Hb_002045_210 |
0.1444851904 |
- |
- |
PREDICTED: protein LONGIFOLIA 1 [Jatropha curcas] |
| 7 |
Hb_001431_090 |
0.1466123388 |
transcription factor |
TF Family: DBP |
PREDICTED: probable protein phosphatase 2C 47, partial [Camelina sativa] |
| 8 |
Hb_005890_020 |
0.1514986185 |
- |
- |
putative plant disease resistance family protein [Populus trichocarpa] |
| 9 |
Hb_168918_010 |
0.1533353121 |
- |
- |
pectin acetylesterase, putative [Ricinus communis] |
| 10 |
Hb_001186_100 |
0.1583963352 |
- |
- |
PREDICTED: uncharacterized protein LOC105640806 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000497_250 |
0.1585606712 |
- |
- |
PREDICTED: potassium transporter 5 [Jatropha curcas] |
| 12 |
Hb_005496_040 |
0.1590520069 |
- |
- |
PREDICTED: uncharacterized protein LOC105630864 [Jatropha curcas] |
| 13 |
Hb_000479_020 |
0.1612031496 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000925_150 |
0.1640812418 |
- |
- |
PREDICTED: aluminum-activated malate transporter 8-like [Jatropha curcas] |
| 15 |
Hb_000260_380 |
0.1643001047 |
- |
- |
PREDICTED: cell division control protein 2 homolog C [Jatropha curcas] |
| 16 |
Hb_011501_010 |
0.1647275431 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG5-like [Jatropha curcas] |
| 17 |
Hb_000270_100 |
0.1662811041 |
- |
- |
ATPase 4, plasma membrane-type -like protein [Gossypium arboreum] |
| 18 |
Hb_000107_360 |
0.166414731 |
- |
- |
PREDICTED: xylem cysteine proteinase 1 [Jatropha curcas] |
| 19 |
Hb_028487_060 |
0.168346861 |
- |
- |
PREDICTED: 21 kDa protein-like [Jatropha curcas] |
| 20 |
Hb_001514_030 |
0.1685092859 |
- |
- |
PREDICTED: cyclin-D3-3-like [Jatropha curcas] |