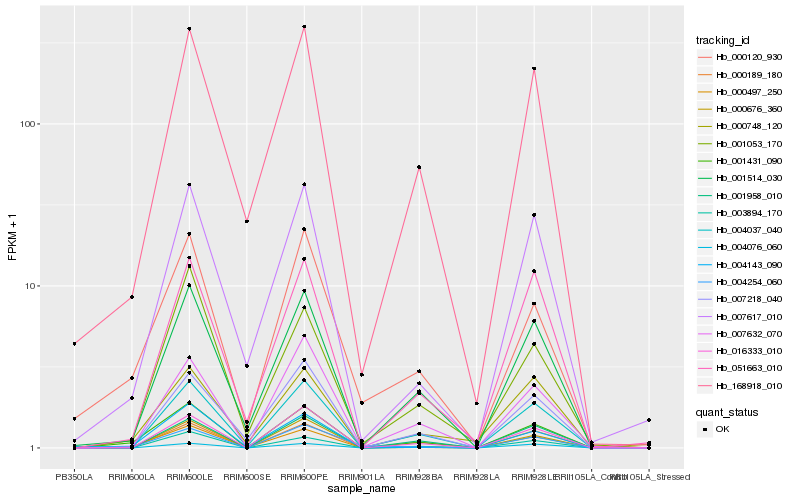
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001431_090 |
0.0 |
transcription factor |
TF Family: DBP |
PREDICTED: probable protein phosphatase 2C 47, partial [Camelina sativa] |
| 2 |
Hb_000497_250 |
0.0880914464 |
- |
- |
PREDICTED: potassium transporter 5 [Jatropha curcas] |
| 3 |
Hb_001514_030 |
0.1345541977 |
- |
- |
PREDICTED: cyclin-D3-3-like [Jatropha curcas] |
| 4 |
Hb_003894_170 |
0.1445608103 |
transcription factor |
TF Family: LOB |
hypothetical protein CICLE_v10021992mg [Citrus clementina] |
| 5 |
Hb_000120_930 |
0.1466123388 |
- |
- |
PREDICTED: G2/mitotic-specific cyclin-2-like isoform X2 [Jatropha curcas] |
| 6 |
Hb_016333_010 |
0.1503597862 |
- |
- |
PREDICTED: probable terpene synthase 13 [Jatropha curcas] |
| 7 |
Hb_168918_010 |
0.1504891512 |
- |
- |
pectin acetylesterase, putative [Ricinus communis] |
| 8 |
Hb_004037_040 |
0.1532190753 |
- |
- |
PREDICTED: putative glucose-6-phosphate 1-epimerase [Jatropha curcas] |
| 9 |
Hb_004143_090 |
0.1542851882 |
- |
- |
PREDICTED: VIN3-like protein 2 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000189_180 |
0.1549506286 |
- |
- |
PREDICTED: calmodulin-like protein 11 [Jatropha curcas] |
| 11 |
Hb_001958_010 |
0.1590403052 |
- |
- |
hypothetical protein CICLE_v100078532mg, partial [Citrus clementina] |
| 12 |
Hb_004254_060 |
0.1590625305 |
- |
- |
PREDICTED: uncharacterized protein LOC105650679 [Jatropha curcas] |
| 13 |
Hb_007617_010 |
0.1613190825 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH63 isoform X1 [Jatropha curcas] |
| 14 |
Hb_007632_070 |
0.1613229 |
- |
- |
kinase, putative [Ricinus communis] |
| 15 |
Hb_007218_040 |
0.1628835614 |
- |
- |
Leucine-rich repeat transmembrane protein kinase protein, putative [Theobroma cacao] |
| 16 |
Hb_000748_120 |
0.1653734805 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL1 [Jatropha curcas] |
| 17 |
Hb_004076_060 |
0.1657628502 |
- |
- |
PREDICTED: UDP-glycosyltransferase 82A1 [Jatropha curcas] |
| 18 |
Hb_001053_170 |
0.1667062412 |
- |
- |
PREDICTED: uncharacterized protein LOC105628694 [Jatropha curcas] |
| 19 |
Hb_051663_010 |
0.1675179966 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 20 |
Hb_000676_360 |
0.1685098541 |
- |
- |
hypothetical protein POPTR_0007s13690g [Populus trichocarpa] |