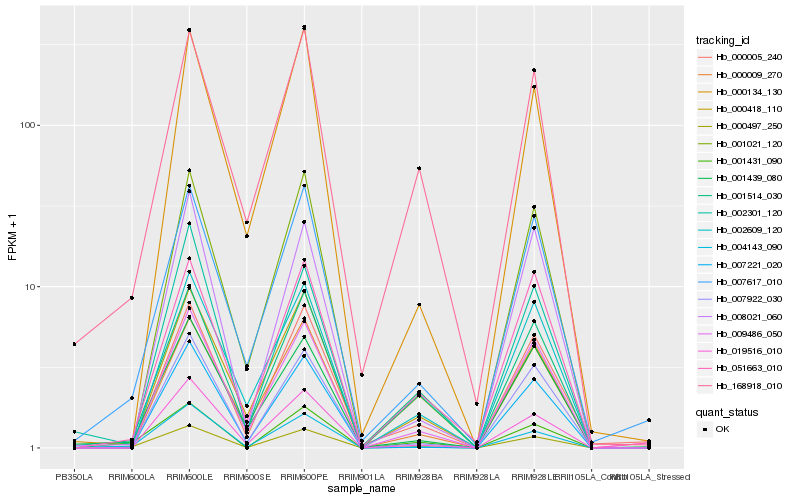
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000497_250 |
0.0 |
- |
- |
PREDICTED: potassium transporter 5 [Jatropha curcas] |
| 2 |
Hb_001431_090 |
0.0880914464 |
transcription factor |
TF Family: DBP |
PREDICTED: probable protein phosphatase 2C 47, partial [Camelina sativa] |
| 3 |
Hb_007617_010 |
0.1008899338 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH63 isoform X1 [Jatropha curcas] |
| 4 |
Hb_004143_090 |
0.1033347424 |
- |
- |
PREDICTED: VIN3-like protein 2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001514_030 |
0.1093211481 |
- |
- |
PREDICTED: cyclin-D3-3-like [Jatropha curcas] |
| 6 |
Hb_019516_010 |
0.1129002048 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 11-like isoform X2 [Populus euphratica] |
| 7 |
Hb_007922_030 |
0.1155968592 |
- |
- |
PREDICTED: uncharacterized protein LOC105111213 [Populus euphratica] |
| 8 |
Hb_001439_080 |
0.1173386527 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 9 |
Hb_001021_120 |
0.1179193973 |
transcription factor |
TF Family: NF-YB |
PREDICTED: nuclear transcription factor Y subunit B-4 [Jatropha curcas] |
| 10 |
Hb_009486_050 |
0.1213256063 |
- |
- |
PREDICTED: cyclin-U4-1 [Jatropha curcas] |
| 11 |
Hb_168918_010 |
0.1236023261 |
- |
- |
pectin acetylesterase, putative [Ricinus communis] |
| 12 |
Hb_000418_110 |
0.12490461 |
- |
- |
PREDICTED: protein kinase PINOID [Jatropha curcas] |
| 13 |
Hb_051663_010 |
0.1257621298 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 14 |
Hb_000009_270 |
0.1265985445 |
- |
- |
Calmodulin, putative [Ricinus communis] |
| 15 |
Hb_007221_020 |
0.1270238411 |
- |
- |
FAD-binding domain-containing family protein [Populus trichocarpa] |
| 16 |
Hb_000005_240 |
0.1281371776 |
- |
- |
Shugoshin-1, putative [Ricinus communis] |
| 17 |
Hb_000134_130 |
0.1293907206 |
- |
- |
PREDICTED: uncharacterized protein LOC105628670 [Jatropha curcas] |
| 18 |
Hb_008021_060 |
0.1296537123 |
- |
- |
Squalene monooxygenase, putative [Ricinus communis] |
| 19 |
Hb_002609_120 |
0.1297550115 |
- |
- |
hypothetical protein POPTR_0001s25660g [Populus trichocarpa] |
| 20 |
Hb_002301_120 |
0.1297816741 |
- |
- |
PREDICTED: uncharacterized protein LOC105649581 [Jatropha curcas] |