| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
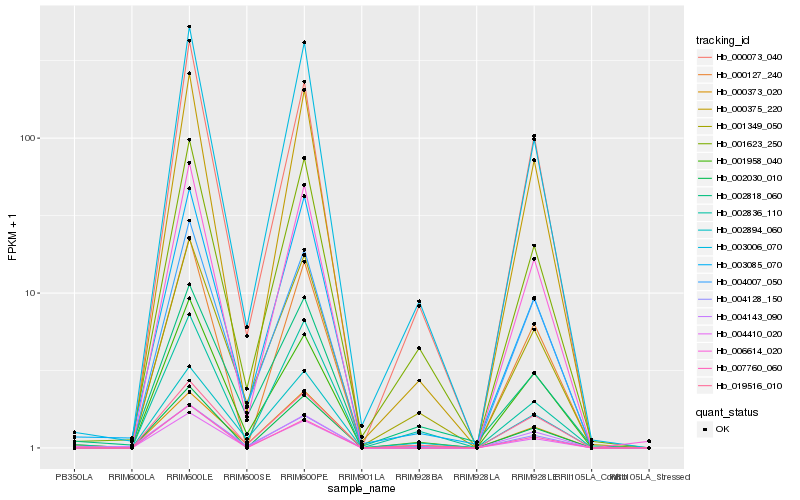
Hb_004410_020 |
0.0 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 2 |
Hb_000373_020 |
0.0876033414 |
- |
- |
PREDICTED: beta-adaptin-like protein A [Jatropha curcas] |
| 3 |
Hb_003006_070 |
0.0903363672 |
- |
- |
aquaporin [Manihot esculenta] |
| 4 |
Hb_003085_070 |
0.0953354525 |
- |
- |
PREDICTED: BURP domain-containing protein 17-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_001623_250 |
0.0992617648 |
- |
- |
PREDICTED: probable pectate lyase 8 [Jatropha curcas] |
| 6 |
Hb_002836_110 |
0.1017397502 |
transcription factor |
TF Family: TUB |
PREDICTED: tubby-like protein 8 [Jatropha curcas] |
| 7 |
Hb_000375_220 |
0.1023885177 |
- |
- |
RecName: Full=2-methylbutanal oxime monooxygenase; AltName: Full=Cytochrome P450 71E7 [Manihot esculenta] |
| 8 |
Hb_004143_090 |
0.1025012508 |
- |
- |
PREDICTED: VIN3-like protein 2 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001349_050 |
0.105107187 |
- |
- |
PREDICTED: aspartic proteinase nepenthesin-1 [Jatropha curcas] |
| 10 |
Hb_006614_020 |
0.1062685375 |
- |
- |
PREDICTED: probable 2-oxoglutarate/Fe(II)-dependent dioxygenase [Jatropha curcas] |
| 11 |
Hb_000073_040 |
0.1096049503 |
- |
- |
hypothetical protein POPTR_0003s21020g, partial [Populus trichocarpa] |
| 12 |
Hb_004128_150 |
0.1104535711 |
- |
- |
hypothetical protein POPTR_0016s08360g [Populus trichocarpa] |
| 13 |
Hb_001958_040 |
0.1159433373 |
- |
- |
PREDICTED: uncharacterized protein LOC105641237 [Jatropha curcas] |
| 14 |
Hb_002818_060 |
0.1187582109 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 15 |
Hb_004007_050 |
0.1188617134 |
transcription factor |
TF Family: GRAS |
PREDICTED: protein SHORT-ROOT [Jatropha curcas] |
| 16 |
Hb_002030_010 |
0.1193187245 |
- |
- |
hypothetical protein JCGZ_18681 [Jatropha curcas] |
| 17 |
Hb_019516_010 |
0.1199269799 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 11-like isoform X2 [Populus euphratica] |
| 18 |
Hb_000127_240 |
0.1201732806 |
- |
- |
SAUR39 [Populus tomentosa] |
| 19 |
Hb_007760_060 |
0.1250727258 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0010s17470g [Populus trichocarpa] |
| 20 |
Hb_002894_060 |
0.1255685812 |
- |
- |
respiratory burst oxidase D [Manihot esculenta] |