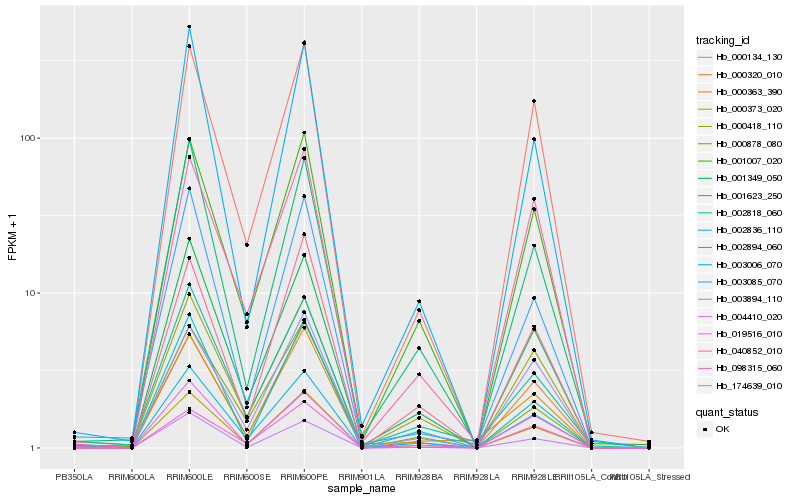
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000373_020 |
0.0 |
- |
- |
PREDICTED: beta-adaptin-like protein A [Jatropha curcas] |
| 2 |
Hb_004410_020 |
0.0876033414 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 3 |
Hb_002894_060 |
0.0959281967 |
- |
- |
respiratory burst oxidase D [Manihot esculenta] |
| 4 |
Hb_001007_020 |
0.097912751 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET1 isoform X2 [Jatropha curcas] |
| 5 |
Hb_002836_110 |
0.1003534408 |
transcription factor |
TF Family: TUB |
PREDICTED: tubby-like protein 8 [Jatropha curcas] |
| 6 |
Hb_001349_050 |
0.1004814122 |
- |
- |
PREDICTED: aspartic proteinase nepenthesin-1 [Jatropha curcas] |
| 7 |
Hb_000320_010 |
0.1027142441 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002818_060 |
0.1040505967 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 9 |
Hb_000363_390 |
0.1086179255 |
- |
- |
Benzoate carboxyl methyltransferase, putative [Ricinus communis] |
| 10 |
Hb_000134_130 |
0.1134000978 |
- |
- |
PREDICTED: uncharacterized protein LOC105628670 [Jatropha curcas] |
| 11 |
Hb_000418_110 |
0.1135075093 |
- |
- |
PREDICTED: protein kinase PINOID [Jatropha curcas] |
| 12 |
Hb_003085_070 |
0.1150303321 |
- |
- |
PREDICTED: BURP domain-containing protein 17-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_174639_010 |
0.115354162 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 14 |
Hb_001623_250 |
0.1177265292 |
- |
- |
PREDICTED: probable pectate lyase 8 [Jatropha curcas] |
| 15 |
Hb_003006_070 |
0.1223010061 |
- |
- |
aquaporin [Manihot esculenta] |
| 16 |
Hb_019516_010 |
0.1230596455 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 11-like isoform X2 [Populus euphratica] |
| 17 |
Hb_003894_110 |
0.1272903048 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 4 [Jatropha curcas] |
| 18 |
Hb_040852_010 |
0.1274996039 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000878_080 |
0.1283030297 |
- |
- |
PREDICTED: uncharacterized protein LOC105635777 [Jatropha curcas] |
| 20 |
Hb_098315_060 |
0.1300223533 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 7-like [Jatropha curcas] |