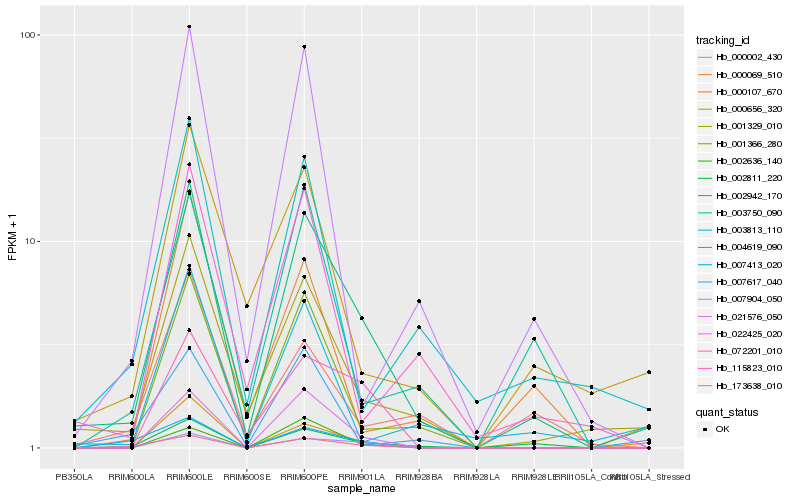
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002942_170 |
0.0 |
- |
- |
acyl carrier protein, putative [Ricinus communis] |
| 2 |
Hb_001329_010 |
0.1598497655 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 3 |
Hb_002811_220 |
0.1693644067 |
transcription factor |
TF Family: Trihelix |
transcription factor, putative [Ricinus communis] |
| 4 |
Hb_022425_020 |
0.180848124 |
- |
- |
- |
| 5 |
Hb_173638_010 |
0.185664066 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 6 |
Hb_007904_050 |
0.1937490098 |
- |
- |
hypothetical protein JCGZ_17883 [Jatropha curcas] |
| 7 |
Hb_001366_280 |
0.1966527864 |
- |
- |
PREDICTED: uncharacterized protein LOC105642732 [Jatropha curcas] |
| 8 |
Hb_115823_010 |
0.2087365811 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000107_670 |
0.215477869 |
transcription factor |
TF Family: MIKC |
floral homeotic protein AGAMOUS [Jatropha curcas] |
| 10 |
Hb_004619_090 |
0.2204986951 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_021576_050 |
0.2208288178 |
- |
- |
PREDICTED: early nodulin-like protein 1 [Jatropha curcas] |
| 12 |
Hb_000656_320 |
0.2215321323 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit gamma 3-like [Jatropha curcas] |
| 13 |
Hb_000069_510 |
0.2271465467 |
- |
- |
PREDICTED: histone H3-like centromeric protein HTR12 [Jatropha curcas] |
| 14 |
Hb_003813_110 |
0.2282099959 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_072201_010 |
0.2282420384 |
- |
- |
PREDICTED: cell division control protein 2 homolog C [Jatropha curcas] |
| 16 |
Hb_007413_020 |
0.2302561041 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_007617_040 |
0.230639421 |
- |
- |
PREDICTED: syntaxin-112-like [Jatropha curcas] |
| 18 |
Hb_002636_140 |
0.2313472298 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g71691-like [Populus euphratica] |
| 19 |
Hb_000002_430 |
0.2328983435 |
- |
- |
PREDICTED: FHA domain-containing protein At4g14490 [Populus euphratica] |
| 20 |
Hb_003750_090 |
0.2335474711 |
- |
- |
BnaC02g34020D [Brassica napus] |