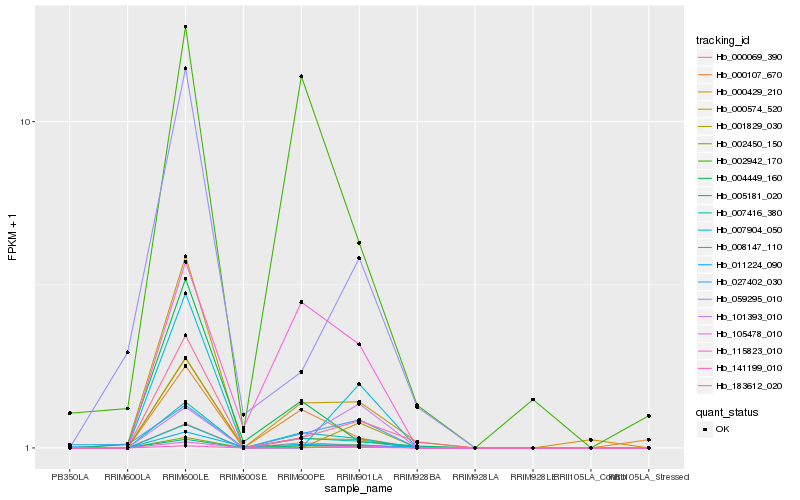
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_141199_010 |
0.0 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 2 |
Hb_005181_020 |
0.0400370413 |
- |
- |
Pectinesterase-2 precursor, putative [Ricinus communis] |
| 3 |
Hb_007416_380 |
0.1136368976 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 3 [Jatropha curcas] |
| 4 |
Hb_000069_390 |
0.1366540704 |
- |
- |
PREDICTED: probable F-box protein At3g61730 [Jatropha curcas] |
| 5 |
Hb_000574_520 |
0.1549703594 |
- |
- |
PREDICTED: uncharacterized protein LOC105634842 isoform X1 [Jatropha curcas] |
| 6 |
Hb_007904_050 |
0.175796702 |
- |
- |
hypothetical protein JCGZ_17883 [Jatropha curcas] |
| 7 |
Hb_183612_020 |
0.1983365835 |
- |
- |
PREDICTED: small heat shock protein, chloroplastic-like [Nelumbo nucifera] |
| 8 |
Hb_002450_150 |
0.2153512142 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 9 |
Hb_115823_010 |
0.2176253672 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_027402_030 |
0.2289348138 |
- |
- |
PREDICTED: probable carboxylesterase 17 [Jatropha curcas] |
| 11 |
Hb_000107_670 |
0.2460665009 |
transcription factor |
TF Family: MIKC |
floral homeotic protein AGAMOUS [Jatropha curcas] |
| 12 |
Hb_101393_010 |
0.2615349177 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_059295_010 |
0.2622013591 |
- |
- |
hypothetical protein CICLE_v10032980mg [Citrus clementina] |
| 14 |
Hb_000429_210 |
0.2700419511 |
transcription factor |
TF Family: ERF |
hypothetical protein RCOM_0089590 [Ricinus communis] |
| 15 |
Hb_002942_170 |
0.2744788945 |
- |
- |
acyl carrier protein, putative [Ricinus communis] |
| 16 |
Hb_008147_110 |
0.2784450443 |
- |
- |
PREDICTED: phenolic glucoside malonyltransferase 1-like [Jatropha curcas] |
| 17 |
Hb_105478_010 |
0.2795374126 |
- |
- |
PREDICTED: uncharacterized protein LOC105762101 [Gossypium raimondii] |
| 18 |
Hb_011224_090 |
0.2796610888 |
- |
- |
PREDICTED: ABC transporter G family member 26 [Jatropha curcas] |
| 19 |
Hb_001829_030 |
0.2812426096 |
- |
- |
PREDICTED: uncharacterized protein LOC105119087 [Populus euphratica] |
| 20 |
Hb_004449_160 |
0.2847097907 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |